Fig. 4 |. Immune cell ecosystem in human breast tissues.
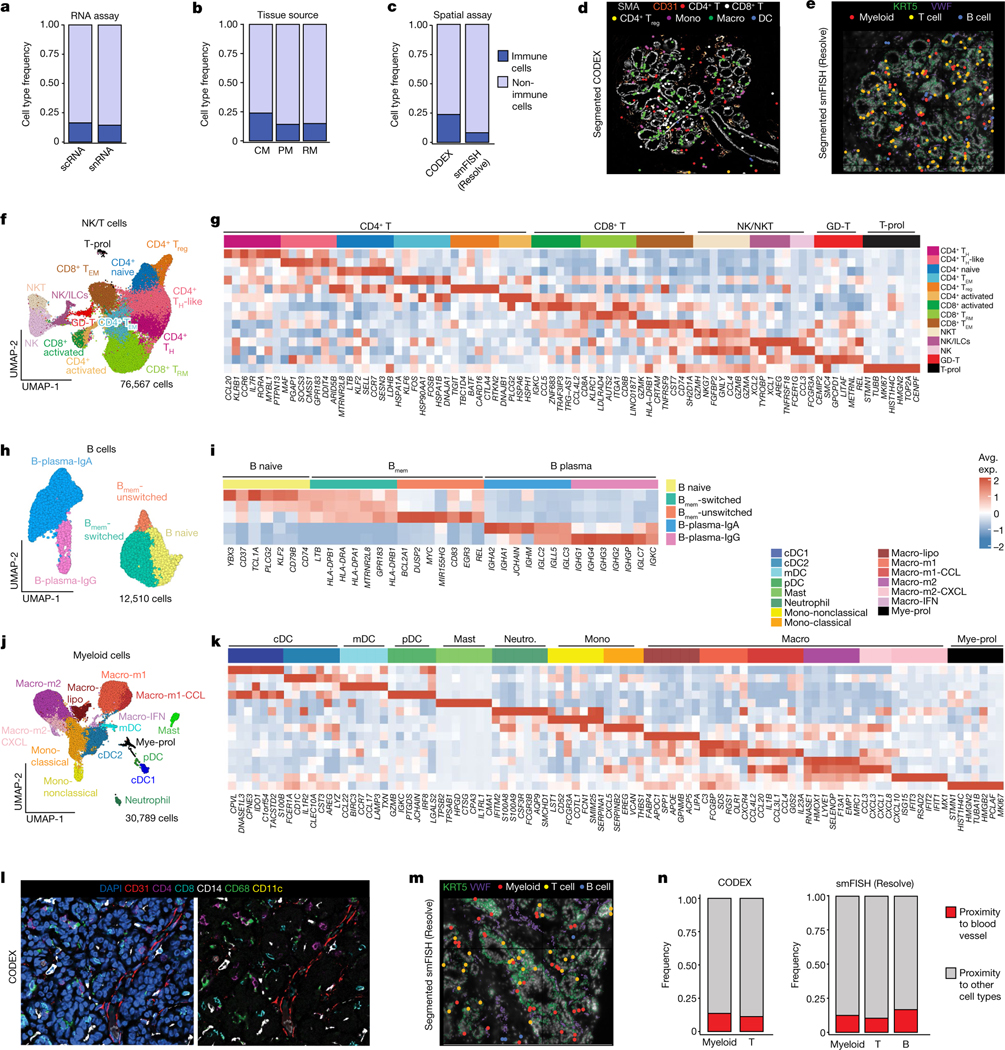
a, Immune and non-immune cell type frequencies in the scRNA-seq and snRNA-seq data. b, Immune and non-immune cell type frequencies by tissue source in the scRNA-seq data. CM, contralateral mastectomies; PM, prophylactic mastectomies; RM, reduction mammoplasties. c, Immune cell type frequencies in the CODEX (n = 8) and smFISH (Resolve) (n = 12) data. d, CODEX data from patient P130 showing a TDLU region with localization of six immune cell types/states. Segmented cells are shown as coloured dots over immunofluorescence staining of SMA (myoepithelial) and CD31 (vessel) for spatial reference. e, smFISH (Resolve) data (P46-S1) showing a TDLU region with localization of three immune cell types. Segmented cells are shown as coloured dots over immunofluorescence staining of KRT5 (basal epithelial) and VWF (endothelial) for spatial reference. f, UMAP representation of 76,567 NK and T cells from scRNA-seq data showing 14 cell states. g, The top genes expressed for each NK and T cell cluster using average values across single cells. h, UMAP representation of 12,510 B cells from scRNA-seq data showing five cell states. Bmem, memory B. i, The top genes expressed for each B cell state using average values across single cells. j, UMAP representation of 30,789 myeloid cells from scRNA-seq data showing of 15 cell types and states. k, The top genes expressed for each myeloid cell cluster using averaged scRNA-seq values. l, CODEX data from patient P130 showing localization of immune cells and a vascular marker (CD31). m, smFISH (Resolve) segmented data (P46-S1) showing localization of immune cells and a vascular marker (VWF). n, The frequency of immune cells that are in the proximity of vascular endothelial cells versus other cell types as determined by neighbourhood analysis of the CODEX and smFISH (Resolve) data.
