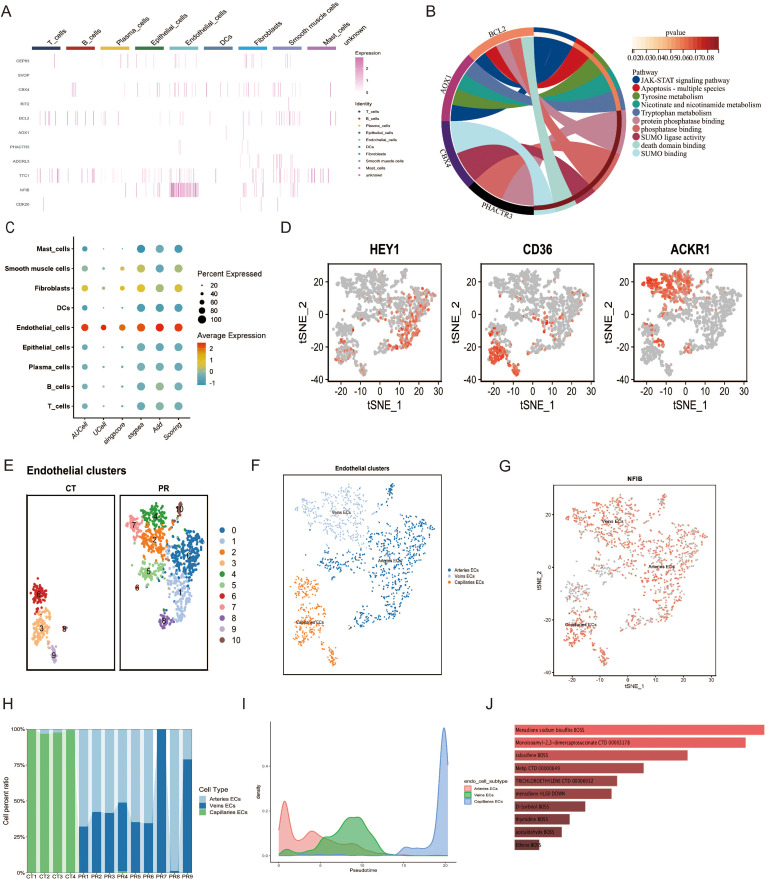
Figure 8.
(A) Heatmap displaying the expression levels of selected genes across various cell types. (B) Chord diagram illustrating the functional pathways associated with selected genes. (C) Dot plot illustrating the scoring of a gene set across various cell types, based on different scoring metrics such as AUCell, UCell, singscore, ssgsea, and Add Scoring. Dot size indicates the percent of cells expressing the gene, and color scale reflects the scoring intensity. (D) t-SNE plots highlighting the expression of marker genes (HEY1, CD36, ACKR1) used for subtyping endothelial cells in single-cell data. Cells expressing each gene are highlighted in red, showing their distribution within the endothelial cell subpopulations. (E) UMAP plot displaying epithelial cell clusters across two sample groups, CT and PR. Each cluster is distinguished by a unique color, representing 11 distinct cell populations. (F) UMAP visualization of endothelial cell subtypes, categorized into arteries ECs (blue), veins ECs (light blue), and capillaries ECs (orange), demonstrating the distinct spatial distribution of these subtypes. (G) Feature plot of the NFIB gene expression across the endothelial cell subtypes in a t-SNE plot. Each dot represents a cell, with color intensity indicating levels of NFIB expression in arteries ECs, veins ECs, and capillaries ECs. (H) Stacked bar chart showing the distribution of endothelial cell subtypes (arteries ECs, veins ECs, capillaries ECs) across different patient samples, displayed as CT1 to CT4 and PR1 to PR9. (I) Density plot representing the pseudotime trajectory analysis of endothelial cell subtypes, illustrating the developmental progression of arteries ECs, veins ECs, and capillaries ECs. (J) Bar chart displaying drugs predicted to target specific genes identified through Mendelian randomization analysis from endothelial cell subtypes, with the bar length indicating the strength of the predicted association based on the analysis.