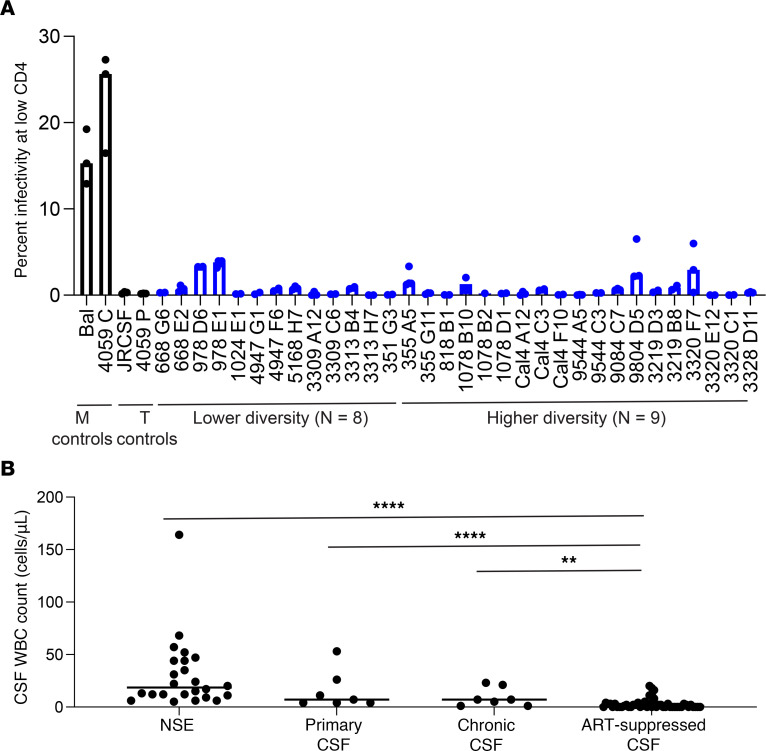
Figure 4. NSE virus is adapted to replicating in CD4+ T cells.
(A) Full-length CSF-derived HIV envs generated by SGA were cloned and used to produce pseudoviruses that were then tested in our Affinofile assay for their ability to facilitate entry into cells with low levels of CD4. None of the NSE envs tested were able to efficiently enter cells with low levels of CD4, but rather required high surface density of CD4 for efficient entry, indicating that the virus was adapted to replicating in T cells. Median relative infectivity values for 2 to 4 biological replicates are plotted with bars, and each individual point is shown with a circle. Representative CSF envs from both the lower diversity group (n = 8) and the higher diversity group (n = 9) were tested (blue bars and circles) as well as macrophage-tropic and T cell–tropic controls (black bars and circles). (B) CSF WBC counts in the NSE cohort and each comparator cohort. Mann-Whitney U tests were performed and found the ART-suppressed cohort (n = 49) had significantly lower CSF WBC counts than participants with NSE (n = 25), untreated primary infection (n = 7), and untreated chronic infection (n = 7): ****P = 0.0002; ****P = 0.0002; **P = 0.0044, respectively. All P values were corrected for multiple comparisons. Median values are shown with horizontal bars.