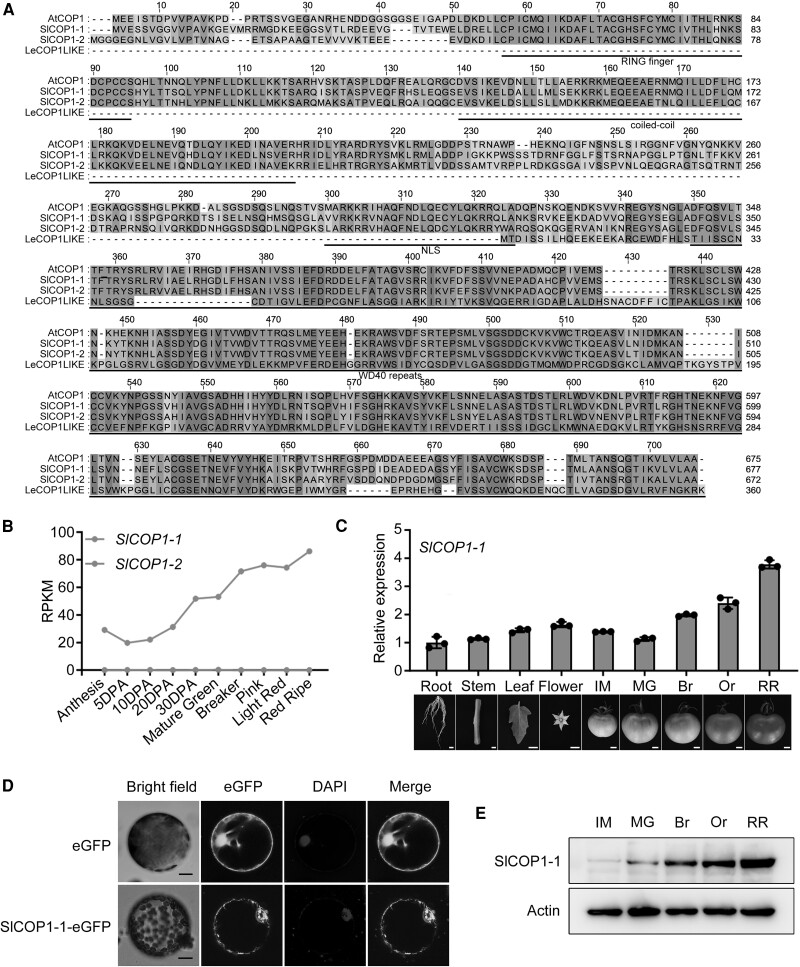
Figure 1.
Identification and characterization of SlCOP1-1. A) Alignment between AtCOP1 and 3 putative tomato COP1 orthologs (SlCOP1-1, SlCOP1-2, LeCOP1LIKE). Functional motifs (RING finger, coiled-coil, WD40 repeats) and the nuclear localization signal (NLS) are underlined based on the AtCOP1structure. Blue indicates identical residues, yellow indicates differing residues, and shades between blue and yellow represent intermediate similarity. B) Expression profile of SlCOP1-1 in tomato cv. Heinz. Data, based on 2 biological replicates from the Tomato Expression Atlas (TEA) database. RPKM, reads per kilobase per million mapped reads. DPA, days post anthesis. C) Expression of SlCOP1-1 in the root, stem, leaf, flower, and fruit at various ripening stages in tomato cv. Ailsa Craig, as determined by RT-qPCR. Values represent means ± standard deviation (SD) of 3 independent experiments. Actin was used as an internal control. IM, immature; MG, mature green; Br, breaker; Or, orange; RR, red ripe. D) Subcellular localization of SlCOP1-1. Protoplasts from Nicotiana benthamiana leaves transiently expressing SlCOP1-1-eGFP were observed by confocal microscopy. 4′,6-diamidino-2-phenylindole (DAPI) was used for nuclear staining. Scale bars, 10 μm. E) Western blot analysis of SlCOP1-1 in fruit at various ripening stages. Actin served as the protein loading control.