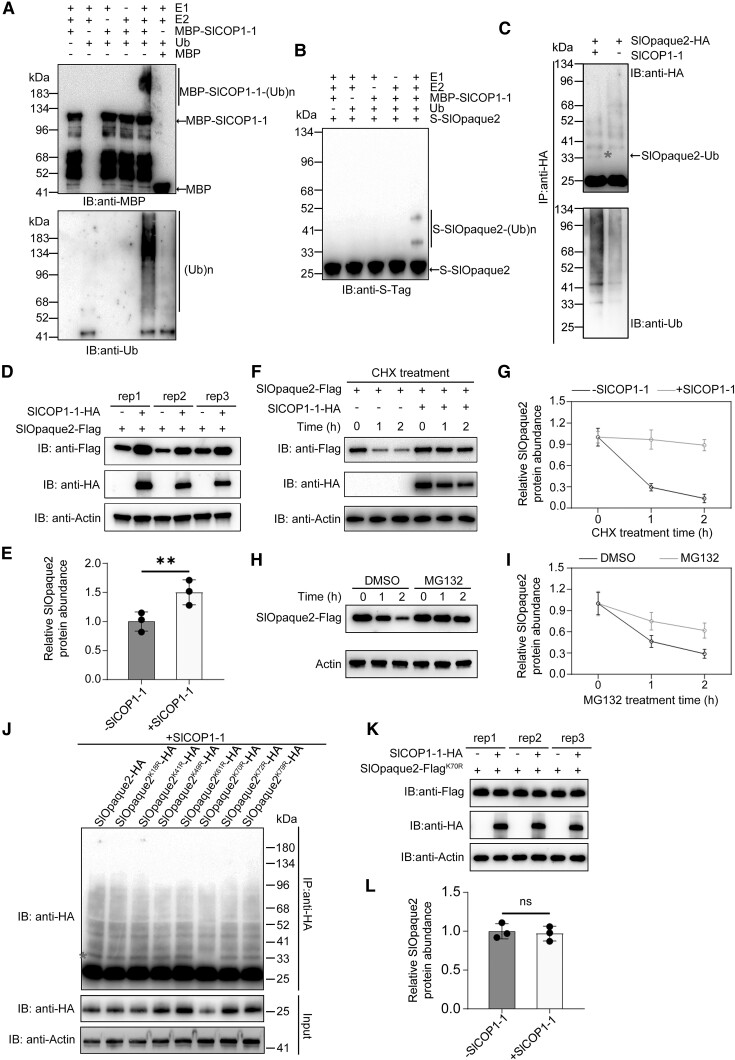
Figure 5.
SlCOP1-1 oligo-ubiquitinates and stabilizes SlOpaque2. A), B)In vitro ubiquitination assay demonstrating SlCOP1-1 as a ubiquitin ligase A) and the ubiquitination of SlOpaque2 by SlCOP1-1 B). Ubiquitination reactions were conducted in the presence (+) or absence (−) of His-tagged ubiquitin (Ub), E1, E2, MBP-tagged SlCOP1-1 (MBP-SlCOP1-1), or S-tagged SlOpaque2 (S-SlOpaque2). The reaction products were subjected to immunoblot analysis using anti-MBP, anti-S-Tag, or anti-Ub antibodies. MBP protein was used as the negative control. (Ub)n, polyubiquitin chain. C) In vivo ubiquitination of SlOpaque2 by SlCOP1-1. SlOpaque2-HA were co-expressed with SlCOP1-1 in Nicotiana benthamiana leaves. Total proteins extracted from transformed leaves were immunoprecipitated with anti-HA beads, followed by immunoblot analysis using anti-HA or anti-Ub antibodies. IB, immunoblot. IP, immunoprecipitation. Red asterisk indicates the mono-ubiquitinated band. D), E) Effect of SlCOP1-1 on the protein stability of SlOpaque2. The SlOpaque2-Flag was expressed in the presence (+) or absence (−) of SlCOP1-1-HA in N. benthamiana leaves. F), G) Degradation rate analysis of SlOpaque2 in the presence (+) or absence (−) of SlCOP1-1. Co-expression of SlOpaque2-Flag with or without SlCOP1-1-HA was performed in N. benthamiana leaves, followed by treatment with translation inhibitor cycloheximide (CHX). H), I) Stability analysis of SlOpaque2. SlOpaque2-Flag was expressed in N. benthamiana leaves followed by treatment with or without the proteasome inhibitor MG132. DMSO, the solvent for MG132, served as a control. For D), F), and H), total protein extracted from the transformed leaves was subjected to immunoblotting analysis using anti-HA or anti-Flag antibodies. Actin was used as the loading control. For E), G), and I), quantification of the immunoblot bands were performed by Image J software. Values are means ± standard deviation (SD) of 3 independent experiments. J) Screening for SlOpaque2 site ubiquitinated by SlCOP1-1 via in vivo ubiquitination assay. All seven lysine (K) sites were individually mutated to arginine (R). The HA-tagged SlOpaque2 variant forms were co-expressed with SlCOP1-1 in N. benthamiana leaves and subjected to ubiquitination analysis as described in C). Red asterisk indicates the mono-ubiquitinated bands. K), L) Protein stability analysis of variant SlOpaque2K70R. The variant SlOpaque2K70R-Flag were co-expressed with SlCOP1-1-HA in N. benthamiana leaves followed by immunoblot analysis K) and quantification L) as described in D) and E). For E) and L), asterisks indicate statistically significant differences (*, P < 0.05, **, P < 0.01, Student's t-test). ns, not significant.