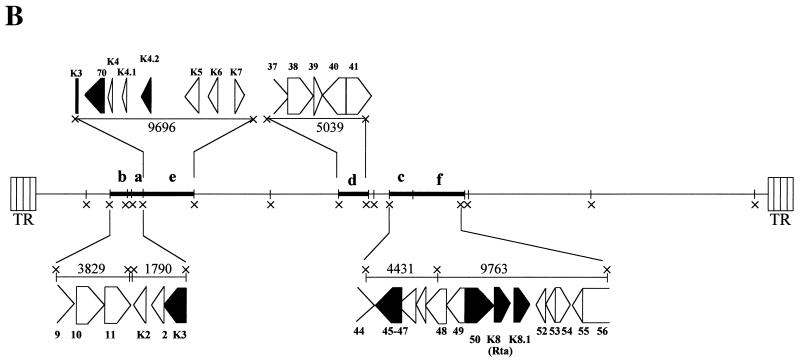
FIG. 2.
Isolation and identification of HHV-8 ORF K3 IE cDNA by SSH. The SSH cDNA was used as a probe for hybridization of HHV-8 genomic DNA digested with the XbaI restriction endonuclease. (A) Southern blot hybridization of HHV-8 genomic DNA with the SSH cDNA probe. Viral genomic DNA extracted from TPA-induced BCBL-1 cell supernatants (1 μg) was extensively digested with XbaI, electrophoresed through a 0.8% agarose gel, transferred to a nylon membrane, and hybridized with the bulk of SSH cDNA labeled with 32P. Six bands (a to f) reacting with the SSH cDNA probe, with sizes ranging from 1.8 to 10 kbp, were identified. The 10-kbp band corresponds to a doublet of similarly sized restriction fragments (e and f) (see panel B). Viral ORFs contained in XbaI restriction fragments a to f (panel B) are indicated on the left side of the autoradiogram. Sequences from ORF K3 are contained in bands a and e. In addition, bands containing larger-sized fragments, possibly corresponding to the largest XbaI restriction fragments or to fragments containing viral terminal repeats, were also detected. HindIII-digested lambda phage DNA was used as a molecular size marker. The positions of the lambda DNA fragments are indicated by dashes; the arrows point to the hybridization bands a to f. The sizes of the DNA fragments are indicated in kilobase pairs. (B) Diagram of HHV-8 XbaI (×) restriction pattern. DNA fragments with sizes corresponding to the hybridization bands reacting with the SSH cDNA probe (a to f) are indicated in bold. The sizes of the reacting fragments (in kilobase pairs) and the encoded HHV-8 ORFs are shown. Restriction fragments a and e contain sequences from ORF K3. ORFs whose sequences are known to be present within HHV-8 IE transcripts are shown in black. Sequences from ORF 70 were determined to be encompassed in the IE ORF K3 transcript (see text for details). TR, terminal repeats.