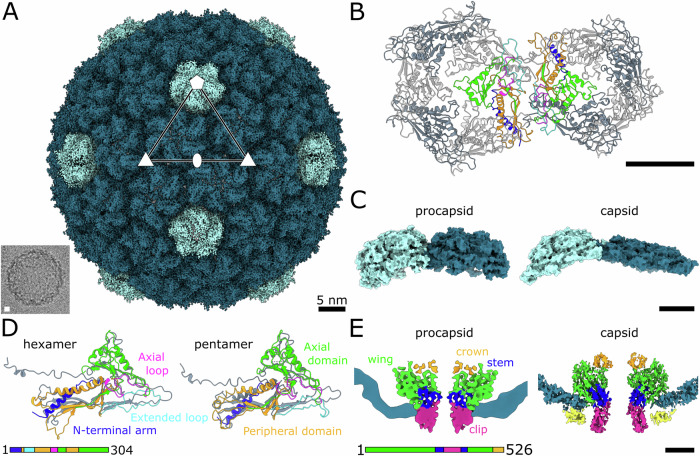
Figure 6. Structure of JBD30 procapsid.
(A) Surface representation of cryo-EM reconstruction of JBD30 procapsid. Pentamers of major capsid protein are highlighted in light blue, hexamers in turquoise. Positions of selected icosahedral five-, three-, and twofold symmetry axes are indicated. The inset shows an electron micrograph of a JBD30 procapsid. (B) Cartoon representation of major capsid proteins forming pentamer and hexamer of procapsid. Two major capsid proteins (one from the hexamer and one from the pentamer) are coloured according to domains as in panel (D). (C) Comparison of pentamers and hexamers from procapsid and capsid. Sideview of the major capsid protein pentamer and hexamer shown as molecular surfaces. (D) Comparison of major capsid proteins from procapsid and capsid. The major capsid proteins from the procapsid are coloured according to domains. Superimposed major capsid proteins from the capsid are shown in grey. Major differences occur in the position of the N-terminal arm and in the width of the extended loop and peripheral domain. (E) Comparison of interactions of portal complex with capsid proteins and in JBD30 procapsid and capsid. Left: Central slice through composite map of portal complex and procapsid. Right: Central slice through asymmetric reconstruction of capsid–portal interface. Density occupied by the major capsid proteins is shown in blue, density of minor capsid proteins is in yellow, density of the portal protein is coloured according to domain. Scale bars 5 nm.