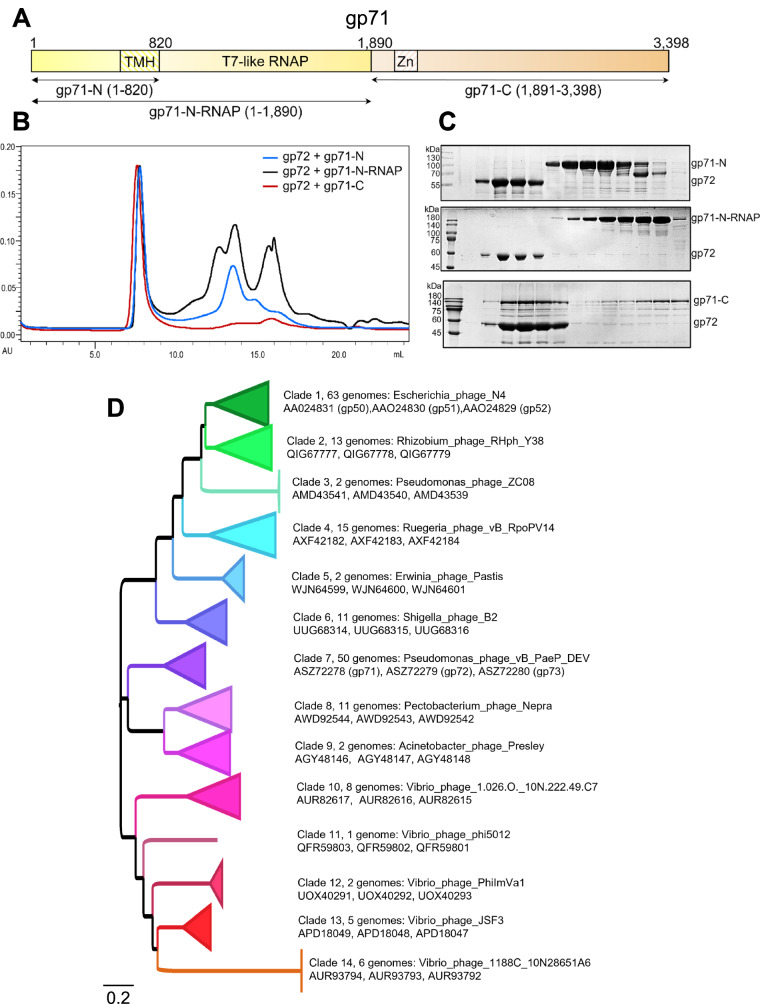
Fig. 8. Domain mapping and conservation of DEV gp71.
A Schematic diagram of gp71 predicted domains. TMH = transmembrane helixes; Zn = zinc-binding domain. B SEC co-migration assay. Overlay of chromatograms obtained by running gp72 with gp71-N (blue line), gp71-N-RNAP (black line), and gp71-C (red line). C SDS-PAGE of fractions eluted from the co-migration assay in panel (B). Gp72 = 59 kDa; gp71-N = 88 kDa, gp71-N-RNAP = 205 kDa and gp71-C = 170 kDa. Each assay was repeated at least three times. Source data are provided as a Source Data file. D Phylogenetic relationships of Schitovirus DEV gp71/ N4 gp50 homologs inferred by maximum-likelihood. 191 homologs of DEV gp71/ N4 gp50 were identified by similarity searches. Various clades have collapsed, and the members of each clade can be found in Supplementary Table S6. Clade refers to groups of Schitoviridae to which the gp71/gp50 homolog was assigned. The longer the branch in the horizontal dimension, the larger the amount of change. The branch length units are amino acid substitutions per position (e.g., the number of amino acid changes divided by the length of the sequence). For each clade, an exemplar genome sequence is indicated, followed by the accession codes for the gp71/gp50 homolog, the inferred gp72/gp51 homolog, and the gp73/gp52 homolog found clustered in the expected sequence on the genome. The tree is arbitrarily rooted.