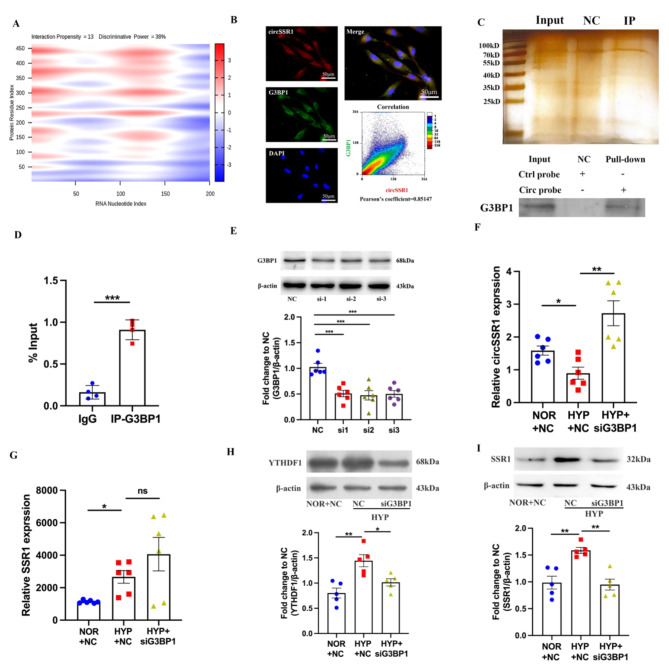
Fig. 6.
G3BP1 can degraded circSSR1 during hypoxia. A, catRAPID predicted heatmap between circSSR1 and G3BP1. B, Immunofluorescence showed G3BP1 can co-localization with circSSR1. C, Fast silver stain kit and RNA pull-down kit were used to verify circSSR1-protein complex pulled down by the circSSR1 probe. D, G3BP1 can combine with circSSR1 proved by RIP experiment (n = 4). E, The small interference RNA (siRNA) efficiency of G3BP1 confirmed by western blot (n = 6). F, qRT-PCR detected the circSSR1 expression interfere with G3BP1 (n = 6). G, qRT-PCR showed G3BP1 not effect on linear SSR1 (n = 6). H-I, Western blot showed that the expression of YTHDF1 and SSR1 protein decreased after interfering with G3BP1 (n = 5). Scale bars, 50 μm. All values are presented as the mean ± SEM. *p<0.05, **p<0.01, ***p<0.001. NOR, normoxia and HYP, hypoxia. NC: negative control