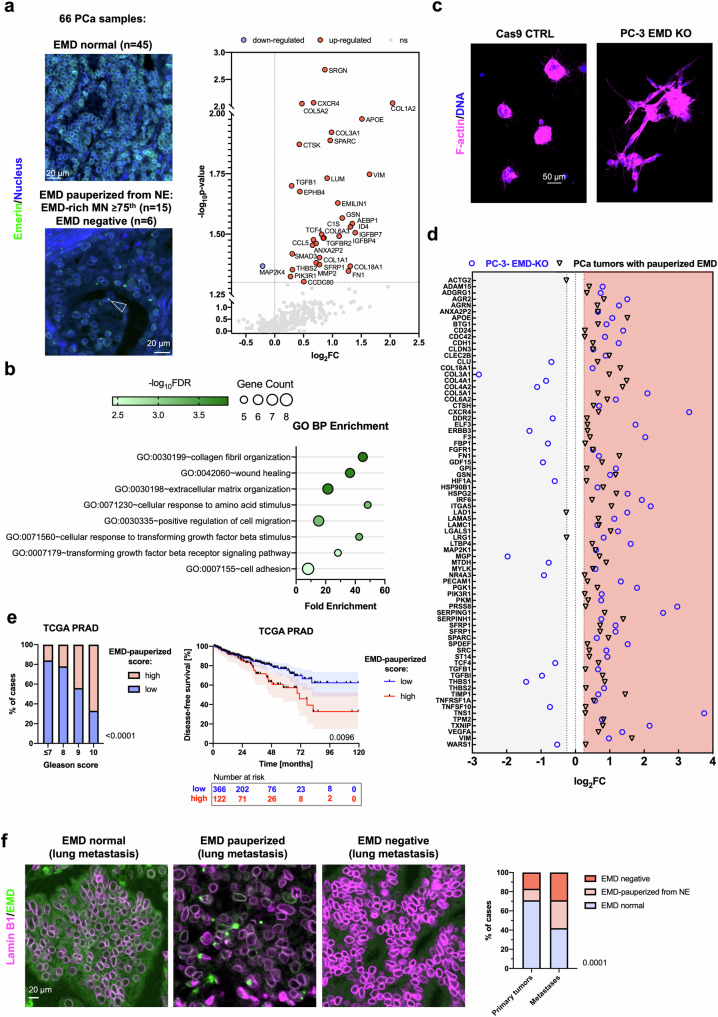
Fig. 5. The transcriptomic signature of PCa cells with Emerin pauperization.
a nCounter analysis of prostate tumors (n = 66) divided into 2 groups based on the pattern of Emerin expression: normal NE expression of Emerin and Emerin pauperization at the NE (presence of Emerin-rich [EMD-rich] MN or negativity for Emerin). The scatter plot shows the differentially expressed genes. b Gene Ontology analysis of upregulated genes in tumors with Emerin pauperization at the NE. c Growth of PC-3 cells with EMD-KO cells under 3D culture conditions. d Forest plot of 75 genes common to both datasets—the nCounter NanoString dataset for prostate tumors and the RNA sequencing dataset for PC-3 cells grown under 3D culture conditions. To facilitate visual representation, the point representing the ACTG2 gene in the EMD-KO group is not shown in the graph (log2FC = 5.861). e Analysis of Emerin pauperization in the TCGA PRAD dataset and its association with the Gleason score and disease-free survival (n = 488). f Distribution of Emerin phenotypes (normal, pauperized, and negative) in prostate primary tumors (n = 278 samples, corresponding to 117 patients) and prostate cancer metastatic lesions (n = 28 samples, corresponding to 14 patients). Bar plot: chi-square test. Kaplan–Meier curve: log–rank test; the error bars indicate the 95% CIs. p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant.