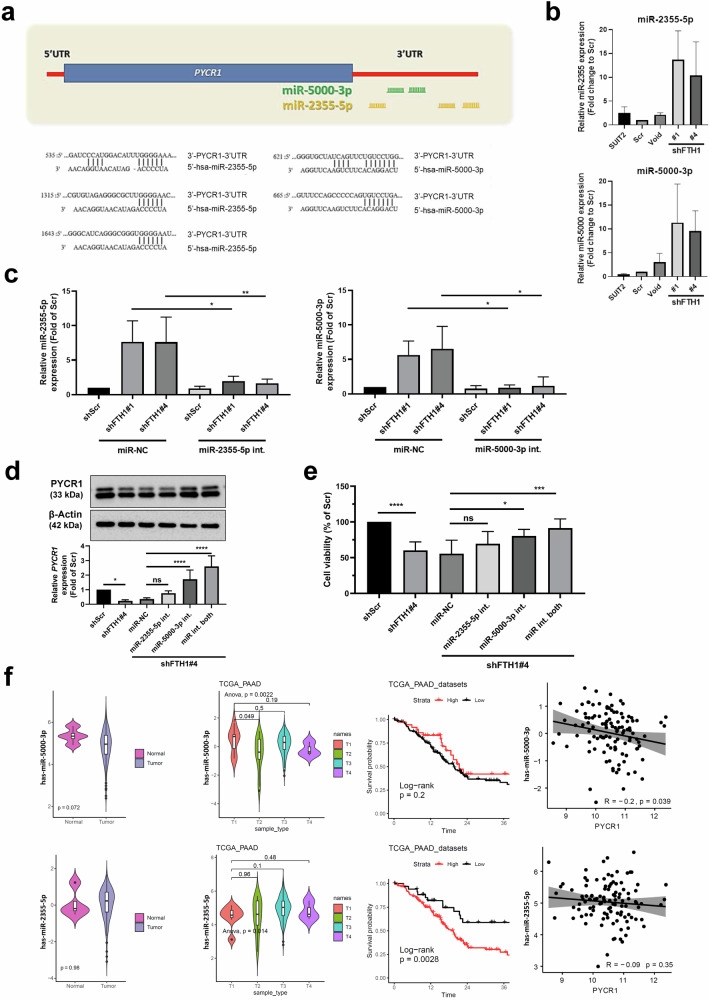
Fig. 7. FTH1 knockdown suppresses PYCR1 expression via miRNA regulation.
a Target prediction via TargetScan revealed that the 3′-UTR sequence of PYCR1 contains putative binding sites for miR-2355-5p and miR-5000-3p. b Expression of miR-2355-5p and miR-5000-3p in the indicated SUIT-2, transfected shCtrl (Scr), and shFTH1 knockdown SUIT-2 (#1 and #4) cells was analyzed using qRT‒PCR. The qRT‒PCR data were normalized to U47 levels in each individual sample, and the bar plot shows the fold changes in Scr expression. The data are expressed as the means ± SEMs from three independent experiments (n = 3). c Differential miRNA expression upon FTH1 knockdown in SUIT-2 cells. Left panel: Relative expression of miR-2355-5p in shScr, shFTH1#1, and shFTH1#4 cells treated with either miR-NC (negative control) or the miR-2355-5p inhibitor (int.). Right panel: Relative expression of miR-5000-3p under the same conditions. The expression was normalized to that in the shScr group, with the bars representing the mean ± SEM. Significance is denoted by asterisks (*p < 0.05, **p < 0.01). d Impact of FTH1 knockdown and miRNA inhibition on PYCR1 protein (upper panel) and mRNA expression (bottom panel) in SUIT-2 cells. β-Actin was used as a loading control for the WBs. Relative PYCR1 mRNA expression in shScr, shFTH1#4, and miR-NC cells and in cells treated with miR-2355-5p or miR-5000-3p inhibitors, both individually and in combination (int. both), normalized to that in shScr-treated cells. e Cell viability of the same groups, expressed as a percentage of the shScr control. Significance is indicated as *p < 0.05, ***p < 0.001, and ****p < 0.0001. f Multipanel analysis of miRNA expression and correlation with survival and PYCR1 levels in PAAD. Top left: Violin plots comparing the expression levels of hsa-miR-5000-3p in normal and tumor tissues, p = 0.072. Top second from left: Violin plots of hsa-miR-5000-3p expression across different tumor stages (T1-T4), with ANOVA p = 0.0022. Top middle: Kaplan‒Meier survival curves stratified by high and low expression of hsa-miR-5000-3p, log-rank p = 0.2. Top right: Scatter plot depicting the inverse correlation between hsa-miR-5000-3p and PYCR1 expression, R = −0.2, p = 0.039. Bottom left: Violin plots showing the expression levels of hsa-miR-2355-5p in normal and tumor tissues, p = 0.98. Bottom second from left: Violin plots of hsa-miR-2355-5p expression across different tumor stages; ANOVA, p = 0.014. Bottom middle: Kaplan‒Meier curves based on hsa-miR-2355-5p expression; log-rank p = 0.0028. Bottom right: Scatter plot showing no significant correlation between hsa-miR-2355-5p and PYCR1 expression, R = −0.09, p = 0.35. The data were derived from the TCGA_PAAD dataset.