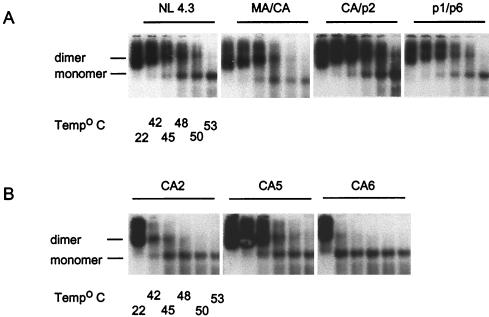
FIG. 3.
Effects of mutations within the primary, secondary, and late cleavage sites on virion RNA dimerization. The impact of mutations within proteolytic processing cleavage sites on genomic RNA dimerization was determined by using melting-curve and electrophoretic analysis of WT and mutant dimers. Virion RNA was resuspended in RNA dimerization buffer and heat denatured for 10 min at the indicated temperatures. Dimers and monomers were electrophoresed in a 1% native agarose gel and probed with an HIV-1 riboprobe, as described in Materials and Methods. (A) RNA dimerization analysis of genomic RNA isolated from WT HIV-1 (NL4.3), CA/p2, MA/CA, and p1/p6 mutant virions. (B) RNA dimerization analysis of genomic RNA isolated from CA2, CA5, and CA6 mutant virions.