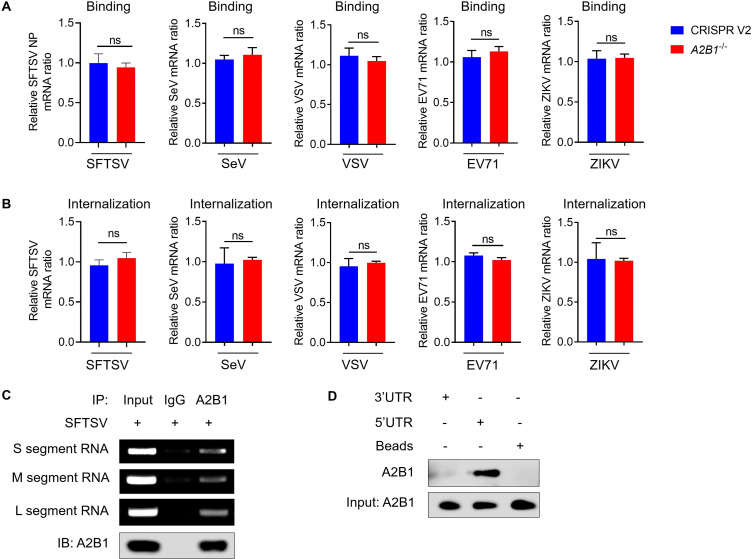
Fig 6.
HnRNP A2B1 interacts with the 5′ UTR of SFTSV RNA. (A) WT and A2B1-/- MEF cells were infected with SFTSV, SeV, VSV, EV71, and ZIKV at an MOI of 10 at 4°C for 1 h, SFTSV NP, SeV, VSV, EV71, and ZIKV mRNA levels were analyzed with RT-qPCR. (B) WT and A2B1-/- MEF cells were infected with SFTSV, SeV, VSV, EV71, and ZIKV at an MOI of 10 at 4°C for 1 h, following incubation at 37°C for 2 h, SFTSV NP, SeV, VSV, EV71, and ZIKV mRNA levels were analyzed with RT-qPCR. (C) MEF cells were infected with SFTSV at an MOI of 10 for 24 h, and the interaction between S, M, or L segment RNA of SFTSV and hnRNP A2B1 was detected with RNA immunoprecipitation (RIP). (D) HEK293T cells were transfected with HA-hnRNP A2B1 plasmids for 24 h, and the interaction between 3′ or 5′ UTR of SFTSV S segment RNA and hnRNP A2B1 was detected with RNA pulldown assay. Data were obtained from three independent experiments (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.