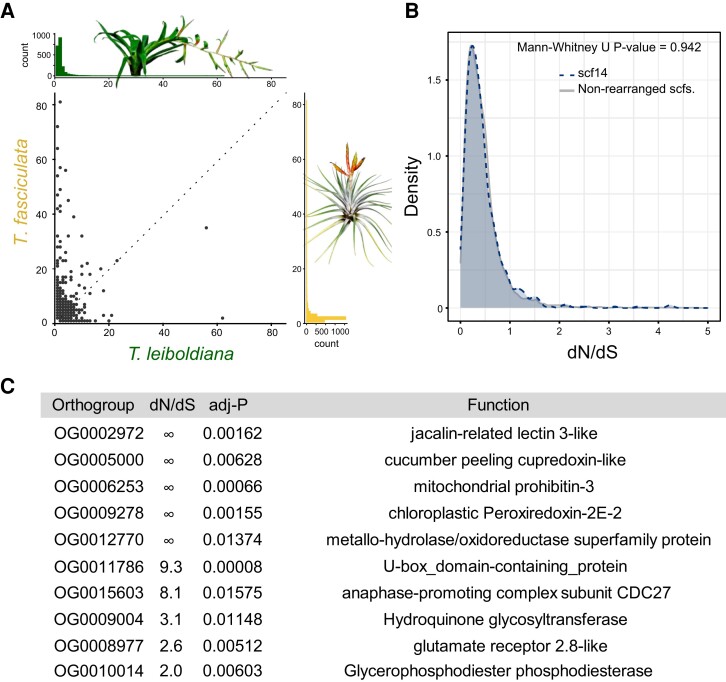
Figure 4.
Analyses of gene family evolution and adaptive sequence evolution linked to large-scale rearrangements between T. fasciculata and T. leiboldiana.A) Scatterplot: composition of per-species gene counts among orthogroups. Upper histogram: distribution of per-orthogroup gene count in T. leiboldiana. Lower histogram: distribution of per-orthogroup gene count in T. fasciculata. B) Density plot showing the distribution of dN/dS values of one-to-one orthologs across non-rearranged scaffolds (gray profile) and scaffold 14 in T. leiboldiana (blue profile), which is the result of a fusion. C) Single-copy orthogroups with significant dN/dS values and their functions. Three uncharacterized genes that are excluded here are detailed in Supplementary Table S6. Infinite dN/dS values correspond to genes with dS = 0 (no synonymous substitutions), an expected situation considering the low divergence of the 2 species. Further explanation about the biological significance of these functions can be found in Supplementary SI Note S7.