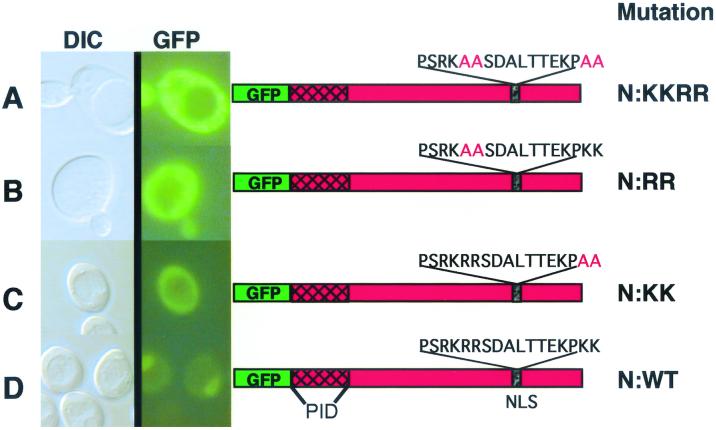
FIG. 3.
Mutational analyses of the carboxy-terminal NLS in the N protein. Shown are differential interference contrast microscopy (DIC) and UV epifluorescence microscopy of yeast cells transformed with plasmids expressing GFP fusions to mutant (A to C) or wild-type (D) N proteins. Note that the GFP–wild-type N fusion (N:WT) localized almost completely to the nucleus, whereas the N-RRKK, N-RR, and N-KK amino acid substitutions of the italicized residues of the bipartite nuclear localization signal (PSRKRRSDALTTEKPKK) resulted in pronounced cytoplasmic fluorescence. The location of the P-interaction domain (PID) of N as defined by two-hybrid experiments is shown. The yeasts were grown and visualized as outlined in the legend to Fig. 2.