Figure 2.
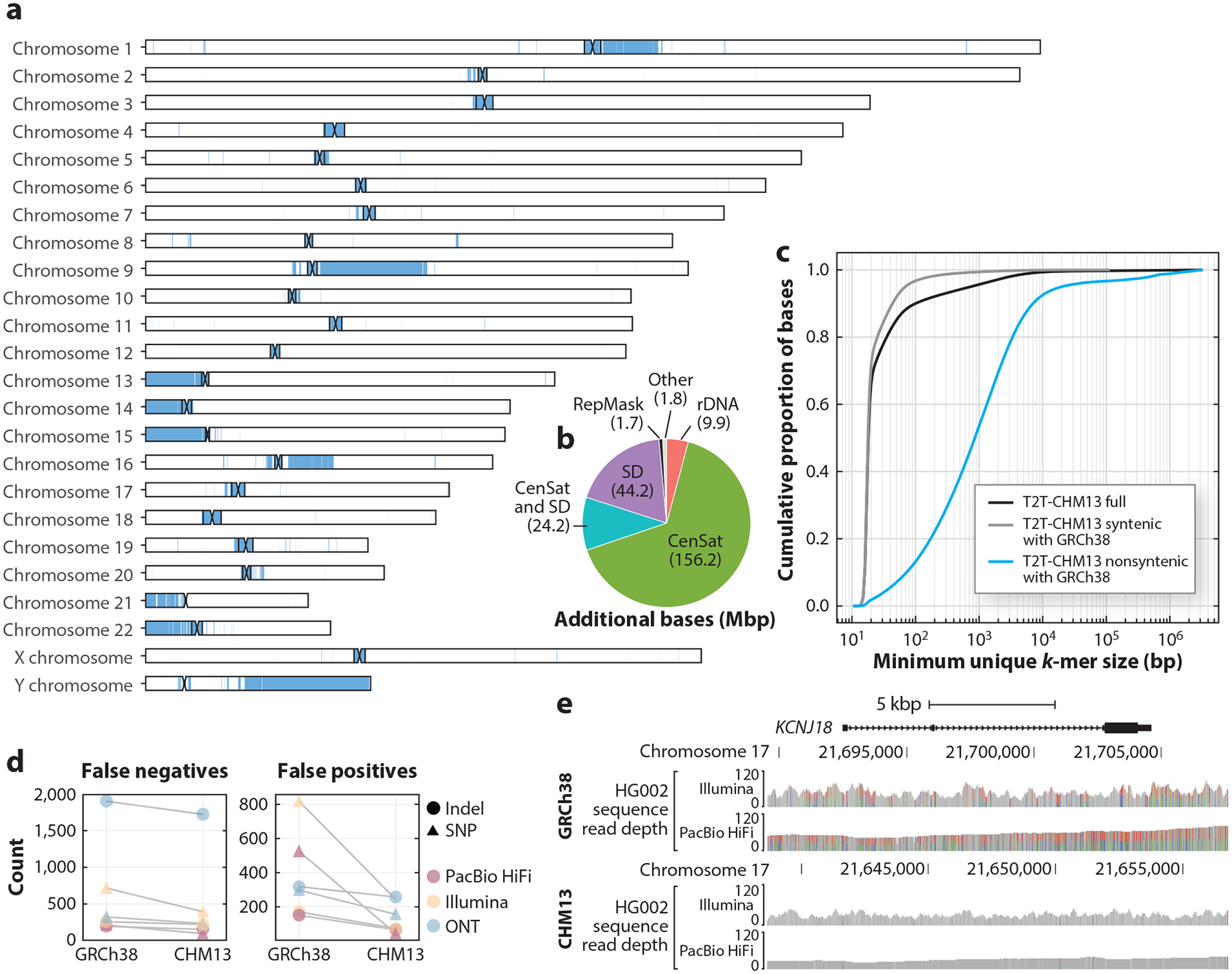
Overview of the first complete human genome assembly. (a) Ideogram of the T2T-CHM13v2.0 genome assembly. Regions of the assembly that are nonsyntenic with GRCh38 based on a whole-genome alignment between the two assemblies are shown in blue. (b) Breakdown of the sequence classes present in the regions of T2T-CHM13 that are nonsyntenic with GRCh38 (Y chromosome not included). (c) Mappability of the T2T-CHM13v2.0 genome based on minimum unique k-mer size, broken down by synteny with GRCh38. At each position in the genome, the minimum unique k-mer size is defined as the minimum number of bases (to the right) necessary to yield a unique sequence that does not appear elsewhere in the genome. Larger sizes imply poor mappability with short sequencing reads. (d) Performance of long- and short-read-based variant identification for a set of challenging medically relevant genes using T2T-CHM13 versus GRCh38. (e) Example of a medically relevant gene exhibiting improved mapping and variant identification using T2T-CHM13. KCNJ18 falls within a collapsed duplicated region in GRCh38, which results in excessive read depth and spurious variants being identified; this is corrected using T2T-CHM13. Abbreviations: CenSat, centromeric satellite; indel, insertion or deletion; ONT, Oxford Nanopore Technologies; RepMask, RepeatMasker; SD, segmental duplication; SNP, single-nucleotide polymorphism; T2T, telomere-to-telomere. Panel b adapted from Reference 110; panels d and e adapted from Reference 5.
