Figure 4.
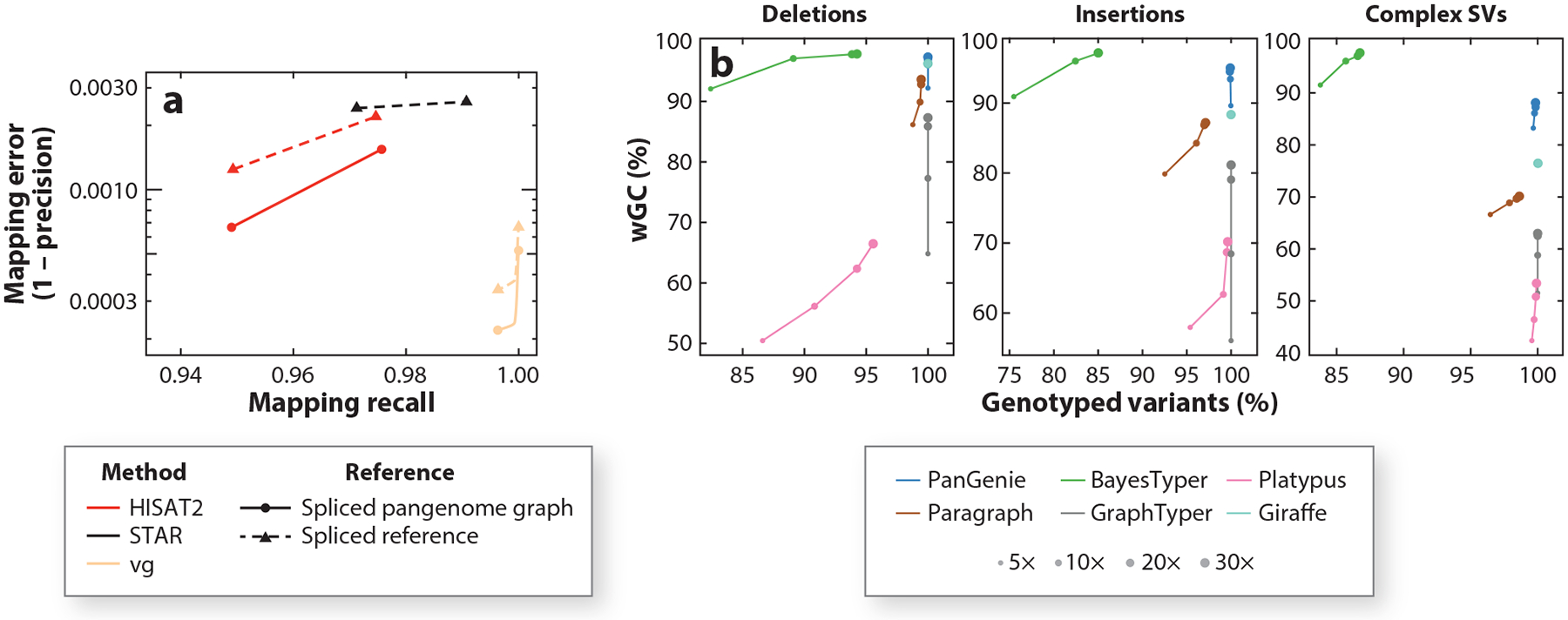
Broader applications of pangenomes. (a) Mapping simulated RNA-sequencing reads to a spliced reference (dashed lines) or spliced pangenome (solid lines). STAR takes only known splicing information into account, while HISAT2 and the vg toolkit also further integrate genetic variants, which results in substantially fewer incorrectly mapped sequencing reads. (b) Genotyping SVs from the HGSVC catalog using different pangenome-based approaches. This panel shows wGC values in nonrepetitive regions, at different coverages (point size), for sample NA12878, which was removed from the catalog for a leave-one-out evaluation. Complex SVs are all variant sites that are not biallelic deletions or insertions. PanGenie is able to genotype the vast majority of SVs accurately. Abbreviations: HGSVC, Human Genome Structural Variation Consortium; HISAT2, Hierarchical Indexing for Spliced Alignment of Transcripts 2; STAR, Spliced Transcripts Alignment to a Reference; SV, structural variant; vg, variation graphs; wGC, weighted genotype concordance. Panel a adapted from Reference 132; panel b adapted from Reference 44 (CC BY 4.0).
