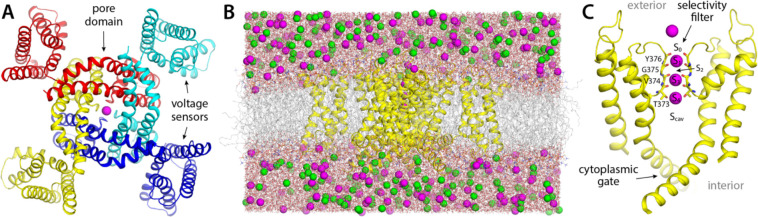
Figure 1. Structure and simulation of the Kv2.1 channel in the activated state.
(A) Structure of Kv2.1 determined by single-particle cryo-electron microscopy in a lipid nanodisc (17), viewed from the cell intracellular side and along the perpendicular to the membrane. The channel is an assembly of four distinct subunits (in colors); the transmembrane ion pore is formed at the center of the assembly, and the voltage sensors are found in the periphery, in a domain-swapped configuration. A K+ ion is shown in magenta, inside the selectivity filter. (B) Simulation system used in this study, comprising the channel (yellow), a phospholipid bilayer (gray), and a 300 mM KCl buLer (magenta, green). The figure depicts the final configuration of the 25-μs MD trajectory described in Figure 2. The total number of atoms is 201,954, some of which are omitted in the figure, for clarity. (C) Close-up of the pore domain and the selectivity filter. Note two protein subunits are omitted, for clarity. The configuration represented is that shown in panel (B); in this configuration, K+ ions are found in sites S1, S3 and S4 within the selectivity filter, while sites S0, S2 and Scav are transiently vacant.