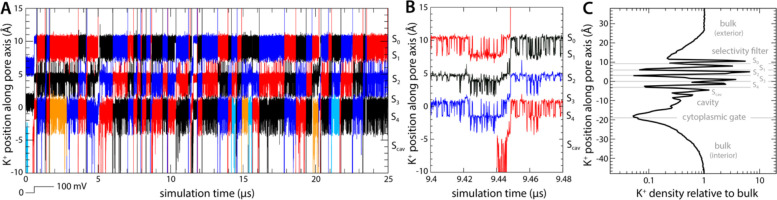
Figure 2. Mechanism of K+ permeation through the Kv2.1 channel.
(A) Time traces of the position of K+ ions along the central axis of the channel as they reach and permeate the selectivity filter towards the extracellular side (black, red and blue). Ions that do not reach the filter are not shown for clarity (those that reach the filter but return to the cytoplasmic side before permeating are shown in orange and cyan). The approximate location of each of the K+ binding sites (S0 through S4, and Scav) along the pore is indicated alongside the plot. (B) Same as (A), for a fragment of the trajectory that illustrates the knock-on mechanism that initiates and completes each of the observed permeation events. (C) Density for K+ ions along the channel axis, relative to the bulk concentration value of 300 mM. Density peaks correspond to each of the K+ binding sites within and adjacent to the selectivity filter. For reference, gray horizontal lines indicate the average position of selected protein atoms: in the selectivity filter these are, from top to bottom, the backbone carbonyl oxygens of residues Y376, G375, V374 and T373, and the sidechain hydroxyl oxygen of T373; and in the cytoplasmic gate, the alpha-carbon of residue P408. All ions in the simulation system contribute to this profile, but only while they reside in a cylindrical volume of diameter equal to 12 Å, centered in and parallel to the channel axis, extending across the whole system.