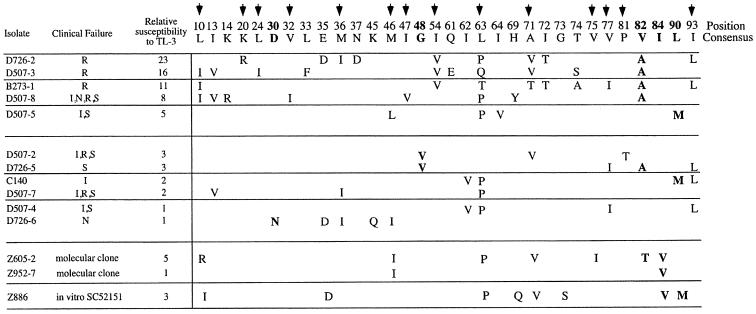
FIG. 1.
TL-3 cross-resistance to and amino acid sequence of protease inhibitor-resistant HIV-1 isolates and isolates obtained from patients failing antiretroviral therapy. The numbered protease consensus amino acid sequence is shown. Bold arrows indicate amino acids associated with resistance when present with other mutations, and amino acids in boldface are active-site mutations. Viral isolates, in the first column, starting with B, C, or D, were obtained from patients experiencing a viral rebound during treatment with the protease inhibitor(s) indicated in the second column. Inhibitors: I, indinavir; N, nelfinavir; S, saquinavir; R, ritonavir. The Z886 isolate was resistant to SC52151. Relative susceptibility to TL-3 is expressed as the relative IC50 compared to the control virus, R8, and values shown are the means from two independent experiments. Significant resistance was defined as a >4-fold-higher IC50 compared to R8.