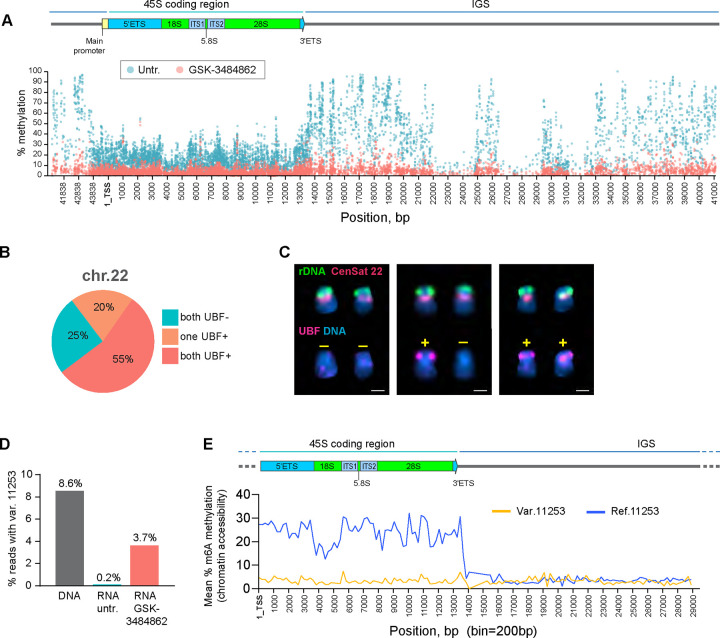
Figure 4. DNMT1 inhibitor restores the activity of silent rDNA arrays in CHM13 cell line.
A. Methyl-sequencing analysis of CHM13 cells grown in the presence of the DNMT1 inhibitor GSK-3484862 for four weeks, with an untreated culture maintained in parallel. The percent methylation of each cytosine base in reads mapped to the rDNA reference sequence across the entire rRNA gene is shown. The DNMT1 inhibitor reduces DNA methylation levels throughout the gene, including the promoter, coding region, and intergenic spacer.
B. UBF immuno-FISH results from forty chromosome spreads of CHM13 cells treated with the DNMT1 inhibitor. The fractions of spreads where one, both, or neither rDNA arrays on chromosome 22 re-gained UBF signal are shown.
C. Examples of chromosome 22 pairs from UBF immuno-FISH experiment show panel in B. The top rows of chromosomes show FISH labeling with rDNA probe (green) and CenSat 22 (red). The bottom rows show corresponding chromosomes with UBF antibody labeling (magenta). DNA was counter-stained with DAPI. UBF status is indicated by +/−. Complete karyograms are provided in Supplementary Figure 6B.
D. Fractions of rRNA reads containing a variant at position 44997. RNA from untreated and GSK-3484862-treated cells was used to create non-ribodepleted libraries. Illumina short sequencing reads were analyzed using the mpileup tool to detect the presence of the variant at position 44997. DNA from the CHM13 cell line was analyzed in parallel. The DNMT1 inhibitor treatment caused re-expression of the silent variant.
E. Fiber-seq mean percent methylation of adenine bases in rDNA-containing reads. Reads containing chromosome 22-specific variant at position 11253 (yellow line) are less methylated in the coding region compared to reads containing the reference base (blue line), indicating less accessible chromatin. Bin size 200 bp.