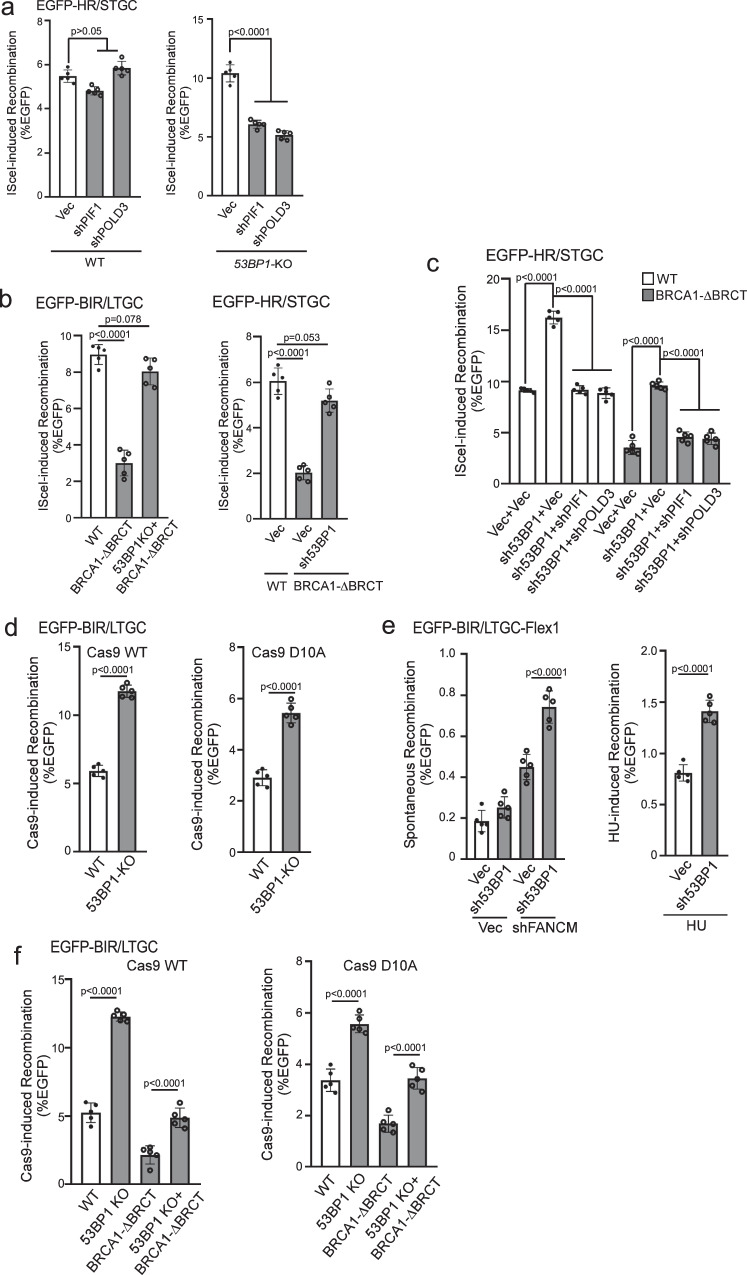
Fig. 3. Hyperrecombination induced by 53BP1 deficiency uses the BIR mechanism.
a U2OS (EGFP-HR/STGC) WT or 53BP1-KO cells stably expressing shRNAs targeting PIF1 or POLD3 were infected with lentiviruses encoding I-SceI, and the percentage of EGFP-positive cells was analyzed by FACS, 5 days post-infection. The expression of PIF1 and POLD3 was examined by qPCR (Supplementary Fig. 3a). (n = 5 replicates). b BIR frequency in U2OS (EGFP-BIR/LTGC) WT, BRCA1-ΔBRCT and 53BP1-KO/BRCA1-ΔBRCT cells (left), and HR frequency in U2OS (EGFP-HR/STGC) WT cells expressing vector, or in BRCA1-ΔBRCT cells expressing 53BP1 shRNA or vector (right), were determined by FACS analysis of EGFP positive cells, 5 days after I-SceI lentiviral infection. (n = 5 replicates). c. U2O (EGFP-HR/STGC) WT and BRCA1-ΔBRCT cells were infected with lentiviruses expressing 53BP1 shRNA or vector, followed by a second round of lentiviral infection with shRNAs targeting PIF1 and POLD3 using vector as a control. HR frequency was determined by FACS analysis of EGFP-positive cells, 5 days after I-SceI lentiviral infection. The expression of indicated proteins was examined by qPCR (Supplementary Fig. 3c). (n = 5 replicates). d BIR frequency was determined in U2OS (EGFP-BIR/LTGC) WT and 53BP1-KO cells by FACS analysis 5 days after lentiviral infection of gRNA/Cas9WT (left) or gRNA/Cas9D10A (right). (n = 5 replicates). e U2OS (EGFP-BIR/LTGC-Flex1) cells expressing 53BP1 shRNA with vector as a control were infected with lentiviruses encoding FANCM shRNA or vector (left), or synchronized to S-phase using double thymidine block followed by HU treatment (2 mM, 24 h, right). BIR frequency was determined by FACS analysis of EGFP positive cells 6 days after. The expression of FANCM was examined by qPCR (Supplementary Fig. 3d). (n = 5 replicates). f BIR frequency was determined by FACS analysis of EGFP positive cells in U2OS (EGFP-BIR/LTGC) WT, 53BP1-KO, BRCA1-ΔBRCT and 53BP1-KO/BRCA1-ΔBRCT cell lines, 5 days after lentiviral infection of gRNA/Cas9WT (left) or gRNA/Cas9D10A (right). (n = 5 replicates). Source data are provided as a Source data file.