Fig. 1 |. Identification of RCAN1 as an age-associated factor in reprogrammed MSNs from longitudinally collected fibroblasts.
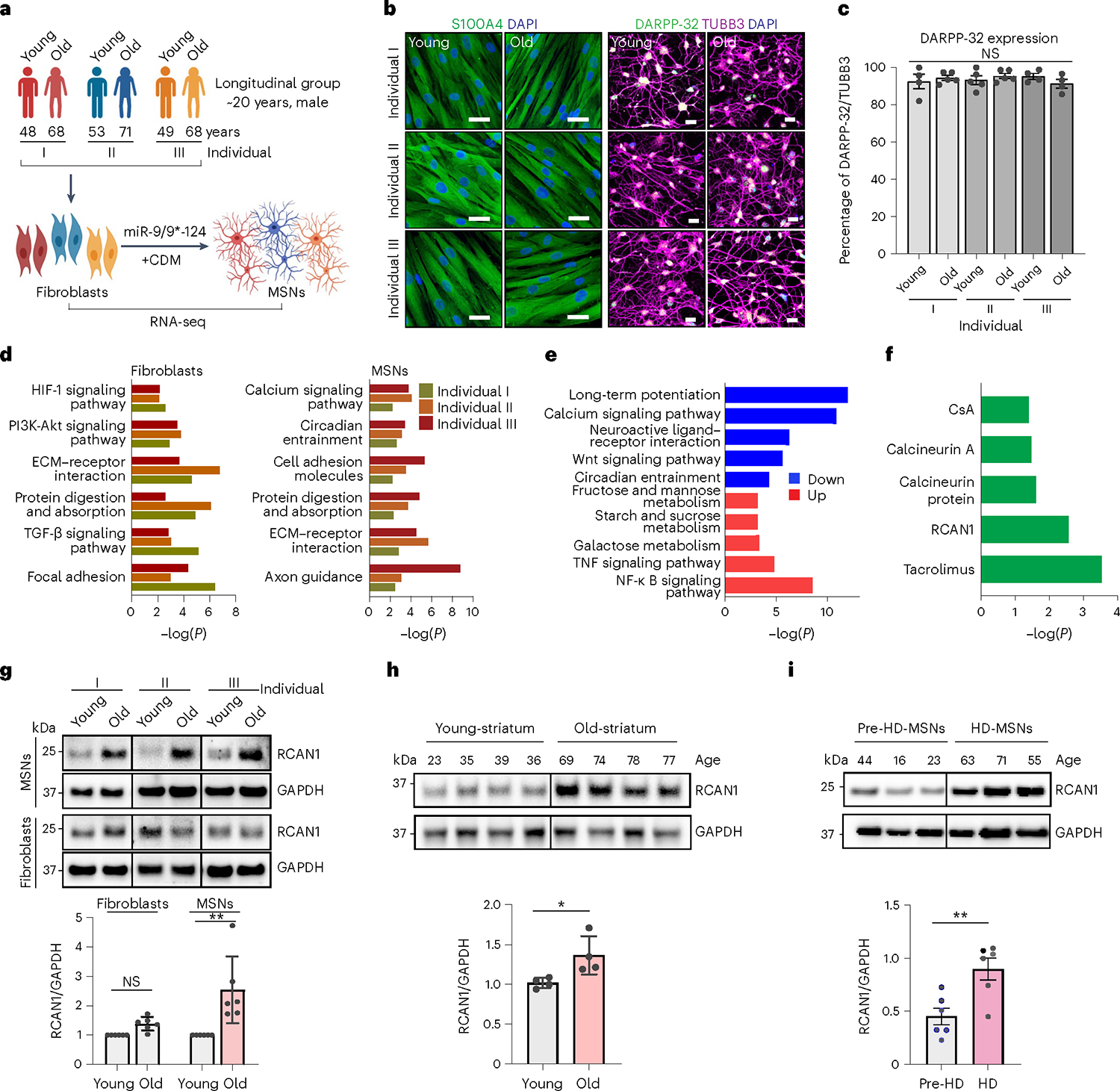
a, Experimental scheme of RNA-seq in fibroblasts and reprogrammed MSNs (young and old) from three independent longitudinal groups from individuals I, II and III. MSNs were reprogrammed by overexpressing miR-9/9* and miR-124 (miR-9/9*−124) as well as MSN-defining TFs CTIP2, DLX1, DLX2 and MYT1L (CDM). b, Representative images of fibroblasts marked by S100A4 (left) and corresponding reprogrammed MSNs marked by DARPP-32 used for RNA-seq. c, Quantification of DARPP-32-positive cells from reprogrammed MSNs from all samples (individual I: young, n = 4; old, n = 5; individual II: young, n = 5; old, n = 5; individual III: young, n = 4; old, n = 4). An average of 300 cells were counted from four or more randomly chosen fields. Scale bars, 20 μm. d, Gene Ontology (GO) enrichment analysis of all DEGs in two replicates of old-fibroblasts (left) and old-MSNs (right) from three independent individuals (FDR < 0.05, | FC | ≥ 1.5). e, GO enrichment analysis of up-/downregulated genes commonly manifested in old-MSNs compared with young-MSNs (FDR < 0.05, | FC | ≥ 1.5). f, Upstream regulator analysis of up-/downregulated genes in old-MSNs in e. g, Representative immunoblotting (top) and quantification (bottom) of RCAN1 in longitudinal MSNs and fibroblasts (young and old) from three independent individuals (n = 6, **P = 0.0077). The quantification is normalized to values from young samples per line. h, Representative immunoblotting (top) and quantification (bottom) of RCAN1 expression in eight human striatum samples from age 23, 35, 39 and 36 (young) and age 69, 74, 78 and 77 (old) individuals (n = 8, *P = 0.0489). i, Representative immunoblotting (top) and quantification (bottom) of RCAN1 expression in three reprogrammed MSNs from presymptomatic patient-derived fibroblasts (44-, 16- and 23-year-old pre-HD-MSN: Pre-HD.42, Pre-HD.45, Pre-HD.40/50) and three reprogrammed MSNs from symptomatic patient-derived fibroblasts (63-, 71- and 55-year-old HD-MSN: HD.47, HD.40, HD.45) (n = 6, **P = 0.0063). Statistical significance was determined using one-way ANOVA with Tukey’s post hoc test (c), two-tailed paired t-test (g) and two-tailed unpaired t-test (h,i); **P < 0.01, *P < 0.05, NS, not significant, and mean ± s.e.m. (c,g,h,i). The sample size (n) corresponds to the number of biologically independent samples (c,g,h,i). DAPI, 4’,6-diamidino-2-phenylindole.
