Figure 5. Novel interaction between CTD of Hsc20 and NBD of Ssq1 is required for the tripartite complex formation.
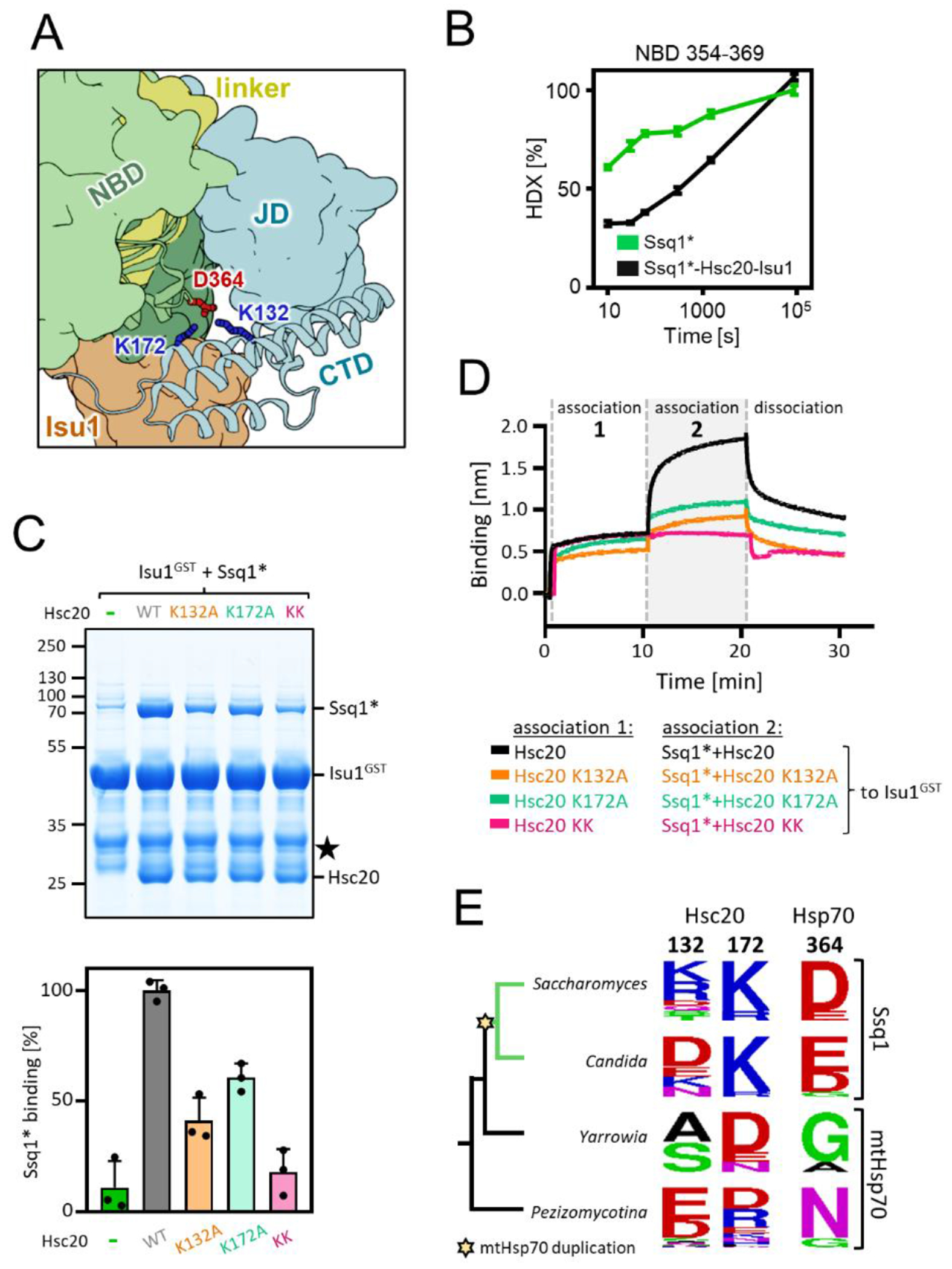
(A) Fragment of the structural model of the tripartite complex showing interaction between K132 and K172 of Hsc20’s CTD and D364 of Ssq1’s NBD- the novel interaction site.
(B) Kinetics of relative deuterium uptake into the D364-containing Ssq1* peptide (354-369). Ssq1* alone (green); tripartite complex (black). (C) (top) Isu1GST (2.5 μM) was incubated with Ssq1* (5 μM) and Hsc20 WT or K132A, K172A or double K132A/K172A (KK) variants (2.5 μM). Glutathione resin was added to pull-down GST and associated proteins, which were then separated by SDS-PAGE and stained with Coomassie blue; entire gel, loading controls and quantification in Figure S12. Molecular weight markers in kDa on left ★ - marks the Isu1GST degradation product. (bottom) Amounts of Ssq1* pulled down were quantified by densitometry and corrected for background binding to GST alone; Ssq1* levels for Isu1GST interacting with Hsc20 and Ssq1* were set at 100%. Error bars represent SD. (D) BLI analysis of Isu1GST interaction with Ssq1* and Hsc20 variants defective in the novel Hsc20-Ssq1 interaction. Association-1, BLI sensors loaded with Isu1GST were placed in solution containing Hsc20 WT or variants K132A, K172A or double K132A/K172A (KK) (1 μM); association-2, the sensors were placed in solution containing Ssq1* (1 μM) and Hsc20 WT or variant (1 μM). Dissociation, the sensors were placed in solution without proteins.
(E) Sequence conservation of Hsc20 and Hsp70 positions involved in the novel Hsc20-Ssq1 interaction in S. cerevisiae across fungi post-duplication species (green branches) in which Hsc20 functions with Ssq1 and non-duplication species (black branches) in which Hsc20 functions with mtHsp70. Star indicates emergence of Ssq1 as result of mtHsp70 gene duplication.
