Fig. 6.
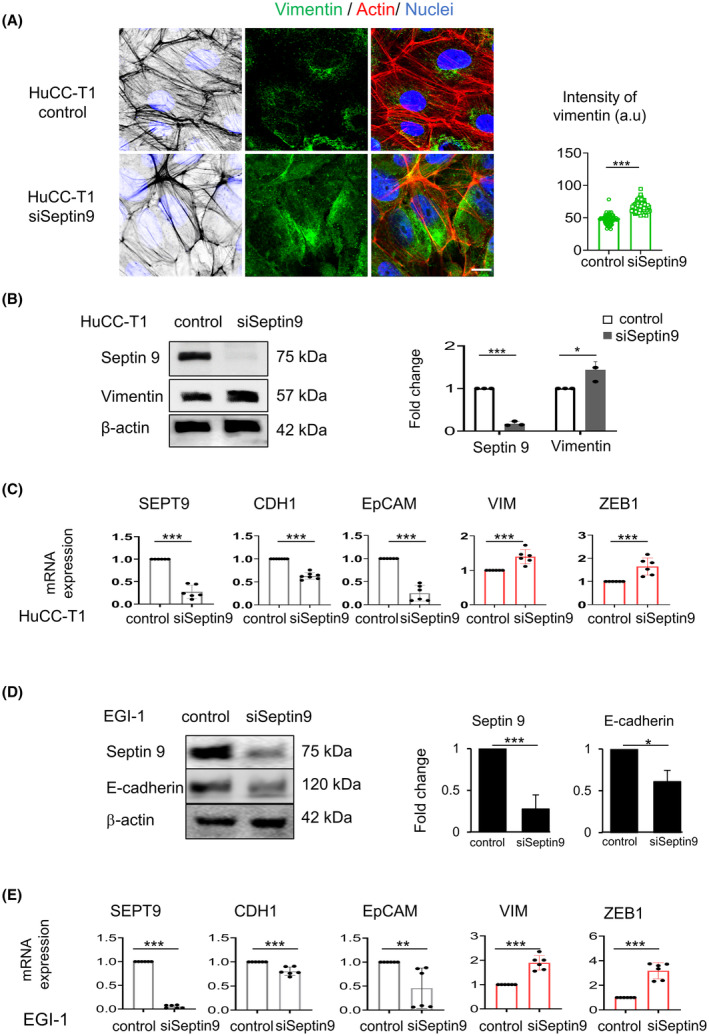
Septin 9 regulates expression of mesenchymal markers in iCCA cells. (A) HuCC‐T1 cells were grown for 24 h and treated with siSeptin 9 or control for 48 h, and then stained for vimentin (green) and actin (red) before analysis using confocal microscopy: scale bar, 10 μm, n = 3. The intensity of vimentin is presented as beside (***P < 0.001, Student's t‐test). (B) HuCC‐T1 cells were treated like (A), then cell lysates were analyzed by immunoblot for septin 9 and vimentin. β‐actin was used as a loading control, n = 3. The fold change of the proteins was shown to the right (*P < 0.05, ***P < 0.001, Student's t‐test). (C) With the same condition as (A), the HuCC‐T1 cells were analyzed for SEPT9, CDH1, EpCAM, VIM, and ZEB1 mRNA expression levels by RT‐qPCR. The bar graphs showed the results, n = 3 (***P < 0.001, Student's t‐test). (D) EGI‐1 cells were treated like (A), then cell lysates were analyzed by immunoblot for septin 9 and E‐cadherin. β‐actin was used as a loading control. The fold change of the proteins was shown to the right n = 3 (*P < 0.05, ***P < 0.001, Student's t‐test). (E) With the same condition as (D), the EGI‐1 cells were analyzed for SEPT9, CDH1, EpCAM, VIM, and ZEB1 mRNA expression levels by RT‐qPCR, n = 3. The bar graphs showed the results (**P < 0.01, ***P < 0.001, Student's t‐test). All data represented mean ± SEM (Student's t‐test). (iCCA, Intrahepatic cholangiocarcinoma; RT‐qPCR, reverse transcription and real‐time PCR analysis; VIM, vimentin gene; CDH1, E‐cadherin gene; EpCAM, epithelial cell adhesion molecule; siSeptin9, siRNA target SEPT9; SEPT9, septin 9 gene).
