Fig. 7.
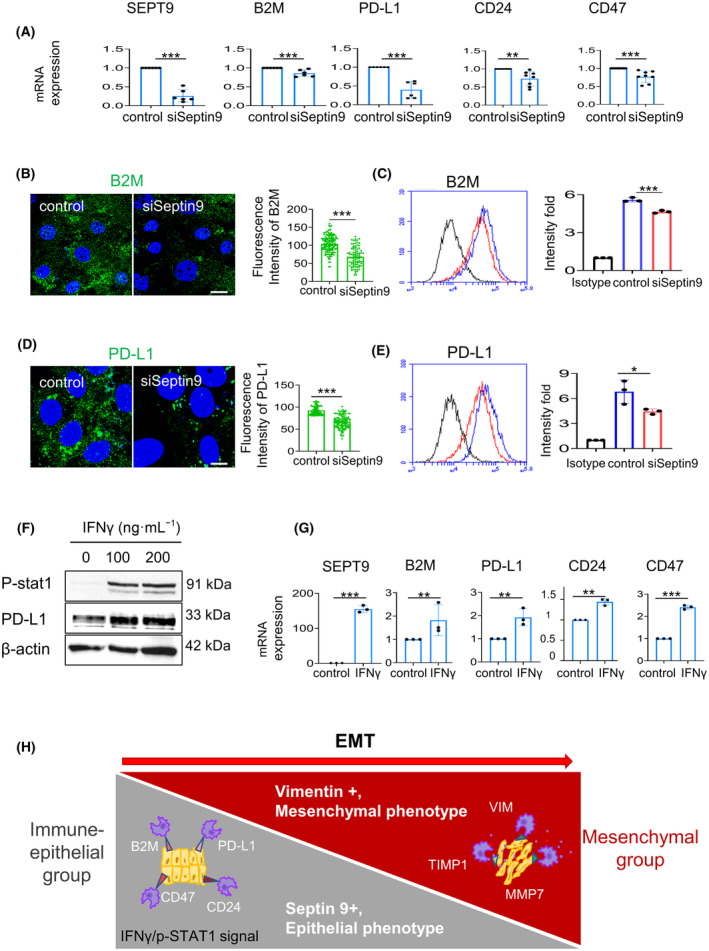
Septin 9 regulates expression of the epithelial‐immune markers in iCCA cells. (A) HuCC‐T1 cells were grown for 24 h and treated with siSeptin 9 or control for 48 h. Then, the cells were analyzed for SEPT9, and ‘don't eat me’ genes mRNA expression by RT‐qPCR (**P < 0.01, ***P < 0.001, Student's t‐test), n = 3. (B, D) HuCC‐T1 cells transfected as in (A) were fixed and then stained for B2M (green) or PD‐L1 (green) before analysis using confocal microscopy: scale bar, 10 μm, n = 3. Next, the quantification of the fluorescence intensity of PD‐L1 or B2M was represented as a histogram beside the image (***P < 0.001, Student's t‐test), n = 3. (C, E) Cells transfected or not with siSeptin9, were stained for B2M or PD‐L1 surface expression, n = 3. The fluorescence intensity was measured by flow cytometry. Representative flow data from three independent experiments are presented as a graph, n = 3. The quantification of the fluorescence intensity fold of B2M or PD‐L1 from different conditions is represented as a histogram (*P < 0.05, ***P < 0.001, Student's t‐test). (F) HuCC‐T1 cells were grown for 24 h and then stimulated with IFN‐ γ in 100 and 200 ng·mL−1 for 24 h, and the cell lysates were analyzed by immunoblot for pSTAT1 and PD‐L1. β‐actin was used as a loading control. The data from a preliminary experiment, n = 2. (G) With the 100 ng·mL−1 IFN‐ γ treatment as (F), HuCC‐T1 cells were collected for SEPT9 and ‘don't eat me’ genes mRNA expression by RT‐qPCR (**P < 0.01, ***P < 0.001, Student's t‐test), n = 3. (H) The schematic graph shows that cytoskeleton septin 9 and vimentin can divide CCA cells into two groups. We termed the septin 9 high expression the ‘Immune‐epithelial group,’ which was featured as epithelial morphology and expressed epithelial markers and the ‘don't eat me’ signal. The IFN‐ γ/p‐STAT1 signal regulated the septin 9 and ‘don't eat me’ signal. The other with high vimentin mesenchymal morphology termed the ‘Mesenchymal group’, contained TIMP1 and MMP7 and mesenchymal markers. All data represented mean ± SEM (Student's t‐test). (iCCA, Intrahepatic cholangiocarcinoma; RT‐qPCR, Reverse Transcription and Real‐time PCR analysis; PD‐L1, programmed cell death ligand 1; B2M, β‐2 macroglobulin subunit of the major histocompatibility class I complex; pSTAT1, phosphorylation of STAT1; IFN‐γ, interferon‐γ; siSeptin9, siRNA target SEPT9).
