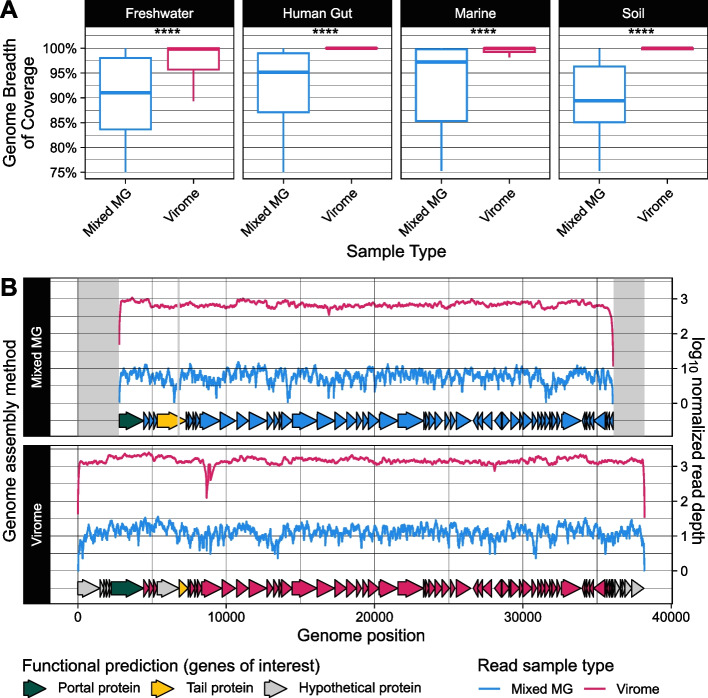
Fig. 4.
Viromes assembled more complete viral genomes than metagenomes. A Distribution of viral genome breadths (percentage of the genome covered by at least one mapped read) in each sample type (viromes and metagenomes). Only viral genomes detected in both sample types and estimated by CheckV [58] to be complete in at least one of the sample types are included. Overall, viral genomes had greater breadths in viromes than metagenomes, indicating that viruses whose genomes were incompletely assembled in metagenomes were more complete in their corresponding virome. Outlier data points are not shown. Significance was inferred by Wilcoxon rank sum test: **** p ≤ 0.0001. B Example of an incomplete metagenome-assembled viral genome that was complete in its corresponding virome. A single-contig, complete viral genome identified from a virome assembly was detected but was incompletely assembled in the sample’s corresponding metagenome. Areas highlighted in gray represent regions in the virome-assembled genome that were absent from the metagenome-assembled genome. Reads yielded from the virome and metagenome of the same sample source were each mapped to both versions of the genome assembly. Arrows along the x-axis represent predicted genes that are colored by the sample type of their genome’s origin, except for a selection of genes of interest that are colored by their functional predictions