Abstract
The ongoing increase in wild boar populations across Europe has fostered human–wildlife conflicts, including the transmission of emerging pathogens with zoonotic importance. Blastocystis is a ubiquitous, faecal-oral transmitted protist that can cause gastrointestinal illnesses and is observed in humans and animals worldwide. The role of wildlife in the epidemiology of Blastocystis is insufficiently understood. Thus, we investigated the occurrence and subtype diversity of Blastocystis in free-ranging wild boars from the Iberian Peninsula using conventional PCR and next-generation amplicon sequencing of a fragment of the ssu RNA gene. A total of 459 wild boar faecal samples were collected across Spain (n = 360) and Portugal (n = 99) between 2014 and 2021. Blastocystis was present in 15.3% (70/459; 95% CI 12.1–18.9) of the wild boars analysed, and its occurrence was significantly higher in Portugal (34.3%, 34/99; 95% CI 25.1–44.6) than in Spain (10.0%, 36/360; 95% CI 7.1–13.6). Seven Blastocystis subtypes (ST5, ST10b, ST13–ST15, ST24b, and ST43) were detected among the surveyed wild boar populations, with greater variability detected in Portuguese samples. ST5 was identified in all the Blastocystis-positive animals, whereas 14.3% of them harboured ST mixed colonisations. Our results demonstrate that Blastocystis ST5 is particularly adapted to infect wild boars. The additional identification of zoonotic STs reinforces the role of wild boars as spreaders of zoonotic infections with public health significance.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13567-024-01385-9.
Keywords: Epidemiology, NGS, subtype diversity wildlife, zoonoses, Spain, Portugal
Introduction
The wild boar (Sus scrofa) is widely distributed in Eurasia, from Europe to the Far East, including Southeast Asia, and extends as far as North Africa, South America, and the USA [1]. In Europe, a remarkable increase in wild boar populations has been recorded in the past four decades as a consequence of its high reproductive rate, supplementary feeding, lack of large predators, land abandonment, shrub encroachment, reduction in the number of human residents in rural areas, intensification of crop production, and changes from harsh to milder winters [2, 3]. Wild boars show a remarkable dispersion ability (more than 45 km on average) [4], colonising an astonishing variety of habitats ranging from the timberline to large cities [5]. Indeed, the wild boar is considered the second most abundant wild ungulate species in Europe, with more than three million individuals estimated [6, 7]. Overabundant and expanding wild boar populations increase human‒wildlife conflicts, including traffic accidents [8, 9], crop damage [10], threats to sensitive areas and species [11, 12], and the transmission of pathogens at the sylvatic‒domestic (including livestock and human) interface [13–15]. The current worldwide distribution and apparent population burgeoning and geographic expansion of wild boars have prompted the consideration of this species as a relevant potential source for emerging animal diseases (some of which are zoonotic), including animal tuberculosis (TB) [16, 17] and re-emerging African swine fever [18], Aujeszky’s disease virus [19], hepatitis E virus, bacteria (e.g., Brucella spp., Erysipelothrix rhusiopathiae), and parasitic infections [13, 14, 20].
The wild boar is one of the most hunted species in Europe, representing a potential source of zoonotic human infections, such as intestinal parasites, that are faecal- orally transmitted indirectly via ingestion of water or food contaminated with faecal material or directly via contact (through carcass handling) with infected animals. The increasing urbanisation of wild boar populations may also raise public health concerns about parasite cross-species transmission at the wild boar–domestic animal–human health interface. Among them, Blastocystis, a member of the Stramenopiles, is a ubiquitous protist that infects/colonises a broad range of human and nonhuman animal hosts [21]. Although Blastocystis is one of the most common microeukaryotes found in the human gastrointestinal tract [22], the clinical significance of Blastocystis is not fully understood. This protist has often been described as an asymptomatic coloniser in large human populations [23]. Furthermore, evidence from recent metagenomic studies suggests that Blastocystis may be part of the healthy gut microbiota in most circumstances [24–27].
Blastocystis is a highly pleomorphic microorganism with wide genetic diversity. On the basis of variability within the small subunit ribosomal RNA (ssu rRNA) gene, Blastocystis can be divided into 44 subtypes (STs) (ST1–ST17, ST21, and ST23–ST48) [28–33]. Among the 16 subtypes reported in humans, ST1 to ST4 are the most common, whereas ST5-ST10, ST12, ST14, ST16, ST23, ST35, and ST41 range from relatively uncommon to rare [34–39]. All other Blastocystis STs have been documented in non-human animal species thus far and are considered to have limited or negligible zoonotic potential. Because of the apparent loose host specificity of multiple Blastocystis STs, surveys investigating the prevalence and molecular diversity of protists from a variety of animal species and geographic origins are of interest to help disentangle the epidemiology and zoonotic potential of Blastocystis STs. This need is particularly evident for wild and domestic ungulate species, for which recent studies have demonstrated complex concomitant colonisation patterns involving up to 18 Blastocystis STs [33, 40–42] and variable associations between age groups and colonisation status [31]. These studies also highlighted the occurrence of cross-species transmission events of uncertain directionality that deserve further investigation.
Of particular interest is assessing intra- and inter-ST discrimination in host infection/colonisation and disease and ascertaining which animal hosts pose a risk to human infection and to what extent. Data on the epidemiology of Blastocystis in wild boar populations are relatively limited (Table 1) [43–57]. In this study, we analysed a large panel of faecal samples from free-ranging wild boars sampled in a broad Iberian geographic range covering Spain and Portugal.
Table 1.
Prevalence and molecular diversity of Blastocystis reported in wild boars (Sus scrofa) globally.
| Country | Population status | Prevalence (no. pos/total) | Detection method | Subtype(s) (n) | Mixed ST’s detected? | References |
|---|---|---|---|---|---|---|
| Brazil | Captive | 13 (10/79) | CM | – | – | [43] |
| Brazil | Captive | 77 (30/39) | PCR, SS | ST1 (3), ST4 (1), ST5 (14), ST8 (1) | No | [44] |
| China | Wild | 0 (0/4) | PCR, SS | – | – | [45] |
| Iran | Wild | 25 (3/12) | CM | – | – | [46] |
| Iran | Wild | 44 (11/25) | CM | – | – | [47] |
| Italy | Wild | 62 (26/42) | PCR, SS, NGS | ST3(1), ST5 (10), ST15 (21) | Yes | [48] |
| Poland | Wild | 50 (1/2) | PCR, SS | ST5 (1) | No | [49] |
| Poland | Captive | 80 (8/10) | PCR, SS | ST5 (8) | No | [50] |
| Portugal | Wild | 29 (42/144) | PCR, SS | ST5 (42) | No | [51] |
| Portugal | Wild | 34 (34/99) | PCR, NGS | ST5 (34), ST10a (1), ST13 (1), ST14 (1), ST15 (1), ST24b (1), ST43 (2) | Yes | This study |
| Slovakia | Captive | 50 (1/2) | PCR, SS | ST12 (1) | No | [52] |
| Slovakia | Wild | ND | PCR, SS | ST15 (4), ST10 (1) | ND | [53] |
| South Korea | Wild | 10 (45/433) | PCR, SS | ST5 (45) | No | [54] |
| Spain | Wild | 0.7 (1/142) | PCR, SS | ST5 (1) | No | [55] |
| Spain | Wild | 10 (36/360) | PCR, NGS | ST5 (22), ST15 (1) | Yes | This study |
| UK | Captive | 50 (1/2) | PCR, SS | ST5 (1) | No | [56] |
| UK | Captive | 50 (2/4) | PCR, SS | ST5 (2) | No | [57] |
Subtypes previously reported in humans (regardless of their true zoonotic potential) are in bold.
CM: Conventional microscopy, ND: Not determined, NGS: Next-generation sequencing, PCR: Polymerase chain reaction, SS: Sanger sequencing.
Materials and methods
Sampling sites and sample collection
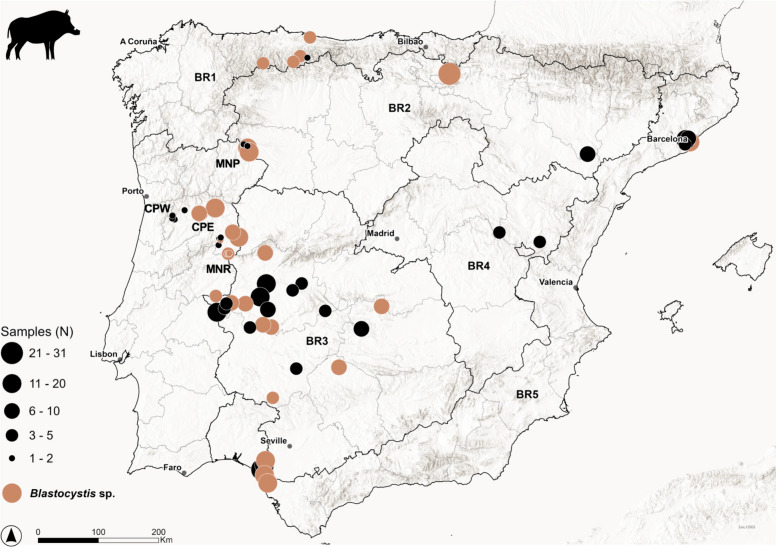
Between autumn 2014 and summer 2021, a retrospective survey was performed in the Iberian Peninsula (Spain and Portugal). Faecal samples from wild boars collected throughout the five bioregions (BRs) of mainland Spain and three comparable BRs in Portugal were used for this purpose (Figure 1). A thorough description of the Spanish BRs can be found elsewhere [58, 59]. The main features of the three adapted Portuguese BRs sampled in the present study and the corresponding locations (MNP—Montesinho Natural Park, CPW—Central Portugal West, CPE—Central Portugal East and MNR—Malcata Nature Reserve) are summarised in Additional file 1 [60].
Figure 1.
Map of the Iberian Peninsula showing the sampled areas in Spain and Portugal and the geographical distribution of Blastocystis detected in wild boar (Sus scrofa). BR2 encompasses Montesinho Natural Park (MNP), BR1 Central Portugal West (CPW), and BR3 Central Portugal East (CPS) and Malcata Nature Reserve (MNR).
The sampling sites included hunting estates or game reserves, natural parks and other classified areas belonging to the European Union’s Natura 2000 Network sites [61]. Faecal samples were collected directly from the rectum of each animal during field necropsies after hunting or from the ground by prospecting several well-distributed transects representative of the different habitats throughout the sampling areas. For the latter case, samples were identified based on the morphology (e.g., content, shape, size) and deposition site by experienced and field-trained personnel. Faecal samples were placed in individually labelled sterile tubes, and collection dates and sites were recorded. Aliquots of these faecal samples were stored at −20 °C by each participating institution responsible for the sampling before being shipped to the Spanish National Centre for Microbiology (SNCM), Majadahonda (Spain), and the Department of Biology & CESAM, University of Aveiro (Portugal), for subsequent molecular analyses.
DNA extraction and purification
Genomic DNA was isolated from approximately 200 mg of each wild boar faecal sample using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, except that samples mixed with InhibitEX buffer were incubated for 10 min at 95 °C. The extracted and purified DNA samples were eluted in 200 µL of PCR-grade water and stored at 4 °C until further molecular analysis. The DNA samples from the extractions carried out at the Department of Biology & CESAM facilities were then shipped to the SNCM for subsequent molecular testing.
Molecular detection and characterisation of Blastocystis using Sanger sequencing
Blastocystis was initially identified via a direct PCR protocol targeting a fragment of the ssu rRNA gene of the parasite [62]. The assay uses the pan-Blastocystis barcode primer pair BhRDr (5ʹ-GAGCTTTTTAACTGCAACAACG-3ʹ) and RD5 (5ʹ-ATCTGGTTGATCCTGCCAGT-3ʹ) to amplify a PCR product of ~ 600 bp. The amplification reactions (25 µL) included 5 µL of template DNA and 0.5 µM of each primer. The amplification conditions consisted of one step at 95 °C for 3 min, followed by 30 cycles of 1 min each at 94 °C, 59 °C and 72 °C, with an additional 2 min final extension at 72 °C.
Amplicons of the expected size were sequenced in both directions by capillary DNA sequencing electrophoresis using BigDye® Terminator chemistry on an ABI PRISM 3130 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA). The obtained consensus sequences were analysed using the Basic Local Alignment Search Tool (BLAST) for Blastocystis confirmation and subtype calling.
Subtype identification using next-generation amplicon sequencing
Subsets of Blastocystis DNA samples whose ssu-PCR amplicons yielded bands of the expected size on agarose gels (regardless of Sanger sequencing confirmation) were shipped to the Environmental Microbial and Food Safety Laboratory, United States Department of Agriculture (Beltsville, Maryland, USA) for subsequent analyses. A next-generation amplicon sequencing (NGS) strategy was used to identify Blastocystis subtypes as previously described [40]. Briefly, a PCR using primers ILMN_Blast505_532F (5ʹ-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGAGGTAGTGACAATAAATC-3ʹ) and ILMN_Blast998_1017R (5ʹ-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTGCTTTCGCACTTGTTCATC-3ʹ (adapter sequences underlined) was used to amplify a fragment of the ssu rRNA gene (ca. 500 bp). These primers were identical to Blast505_532F/Blast998_1017R [63], except that they contained Illumina overhang adapter sequences at the 5′ end. The final libraries were quantified via Qubit fluorometric quantitation (Invitrogen, Carlsbad, CA, USA) before normalisation. A final pooled library concentration of 8 pM with a 20% PhiX control was sequenced using an Illumina MiSeq and a 600 cycle v3 kit (Illumina, San Diego, CA, USA). Paired-end reads were processed and analysed with an in-house pipeline as previously described [40]. The raw FASTQ files were submitted to the NCBI sequence read archive under project number PRJNA1022431. The nucleotide sequences obtained in this study have been deposited in GenBank under the accession numbers OR730909–OR730919, OR730924, OR730933, OR730938, and OR730943–OR730947.
Data analysis
Parasite prevalence was estimated using a binomial test in R software [64], establishing confidence limits with 95% intervals (CI) and χ2 values with the chi-square test function.
Results
Occurrence of Blastocystis
A total of 459 faecal samples were collected across Spain (n = 360) and Portugal (n = 99) between 2014 and 2021 (Additional file 2). Overall, 15.3% (70/459; 95% CI 12.1–18.9) of the faecal samples from the wild boar analysed were confirmed to be positive for Blastocystis by Sanger sequencing and/or next-generation sequencing (NGS). Samples that yielded PCR amplicons of the expected size but could not be confirmed by Sanger sequencing and/or NGS were conservatively considered negative. Wild boars from Portugal presented higher Blastocystis carriage rates (34.3%, 34/99; 95% CI 25.1–44.6) than those from Spain did (10.0%, 36/360; 95% CI 7.1–13.6), and this difference was statistically significant [χ2 (1, n = 459) = 22.1, P < 0.001].
Table 2 shows the distribution of Blastocystis in wild boars from Spain according to the sampling variables considered. The occurrence of protists varied greatly among bioregions [χ2 (4, n = 360) = 23.0, P < 0.001], with animals from BR1 (38.1%) and BR2 (23.1%) having the highest prevalence. All eight animals available from BR4 tested negative for Blastocystis. At the sampling site, wild boars from game reserves were more likely to harbour Blastocystis [χ2 (3, n = 360) = 16.8, P < 0.001]. Blastocystis presence was significantly greater (34.6%) in wild boars sampled in 2014 [χ2 (4, n = 360) = 18.9, P < 0.001], all from the province of Álava in BR2 (Additional file 2).
Table 2.
Prevalence of Blastocystis subtypes in Spanish wild boars (n = 360) according to the bioregion of origin, type of sampling site, and sampling year.
| Variable | Samples (n) | Blastocystis-positive (n)a | Blastocystis-positive (%) | 95% CI (%) | P value | Subtypes detectedb (n) |
|---|---|---|---|---|---|---|
| Bioregion | < 0.001 | |||||
| BR1 | 21 | 8 | 38.1 | 18.1–61.6 | ST5 (8) | |
| BR2 | 39 | 9 | 23.1 | 11.1–39.3 | ST5 (7), ST5/ST15 (1) | |
| BR3 | 150 | 13 | 8.7 | 4.7–14.4 | ST5 (2) | |
| BR4 | 8 | 0 | 0.0 | – | – | |
| BR5 | 142 | 6 | 4.2 | 1.6–9.0 | ST5 (4) | |
| Type of sampling site | < 0.001 | |||||
| Hunting state | 197 | 22 | 11.2 | 7.1–16.4 | ST5 (9), ST5/ST15 (1) | |
| Game reserve | 21 | 8 | 38.1 | 18.1–61.6 | ST5 (8) | |
| Natural protected area | 97 | 4 | 4.1 | 1.1–10.2 | ST5 (4) | |
| Urban/suburban | 45 | 2 | 4.4 | 0.5–15.2 | Not available | |
| Sampling yearc | < 0.001 | |||||
| 2014 | 26 | 9 | 34.6 | 17.2–55.7 | ST5 (7), ST5/ST15 (1) | |
| 2018 | 148 | 6 | 4.1 | 1.5–8.6 | ST5 (4) | |
| 2019 | 52 | 6 | 11.5 | 4.3–23.4 | ST5 (2) | |
| 2020 | 70 | 5 | 7.1 | 2.4–15.9 | Not available | |
| 2021 | 60 | 9 | 15.0 | 7.1–26.6 | ST5 (7) |
95% confidence intervals (95% CI) are included. The values in bold represent statistical significance and subtypes previously reported in humans (regardeless of their true zoonotic potential).
aSamples were considered positive when Blastocystis was identified after Sanger and next-generation sequencing.
bSubtype information is only included for the 22 samples in which next-generation amplicon sequencing was conducted.
cFour samples from unknown sampling years, with one of the samples positive for Blastocystis ST5.
Table 3 shows the distribution of Blastocystis in wild boars from Portugal according to the sampling variables considered. All the investigated animals were from naturally classified areas. Neither the bioregion [χ2 (2, n = 99) = 1.3, P = 0.515] nor the sampling year [χ2 (2, n = 99) = 2.0, P = 0.373] influenced the occurrence of the protist in the investigated wild boar subpopulation.
Table 3.
Prevalence of Blastocystis in Portuguese wild boars (n = 99) according to the bioregion of origin and sampling year.
| Variable | Samples (n) | Blastocystis positive (n)a | Blastocystis positive (%) | 95% CI (%) | P value | Subtypes detectedb (n) |
|---|---|---|---|---|---|---|
| Bioregion | 0.515 | |||||
| BR1 | 12 | 3 | 25.0 | 5.5–57.2 | ST5 (3) | |
| BR2 | 39 | 9 | 23.1 | 11.1–39.3 | ST5 (6), ST5/ST10a (1), ST5/ST14 (1), ST5/ST43 (1) | |
| BR3 | 48 | 22 | 45.8 | 31.4–60.8 | ST5 (18), ST5/ST13 (1), ST5/ST15 (1), ST5/ST24b (1), ST5/ST43 (1) | |
| Sampling year | 0.373 | |||||
| 2019 | 64 | 18 | 28.1 | 17.6–40.8 | ST5 (14), ST5/ST10a (1), ST5/ST14 (1), ST5/ST24b (1), ST5/ST43 (2) | |
| 2020 | 21 | 8 | 38.1 | 18.1–61.6 | ST5 (8) | |
| 2021 | 14 | 8 | 57.1 | 28.9–82.3 | ST5 (6), ST5/ST13 (1), ST5/ST15 (1) |
All the samples were collected from naturally classified areas. 95% confidence intervals (95% CI) are included. BR1 encompasses Central Portugal West (CPW), BR2 Montesinho Natural Park (MNP), and BR3 Central Portugal East (CPE) and Malcata Nature Reserve (MNR). Subtypes previously reported in humans (regardless of their true zoonotic potential) are in bold.
aSamples were considered positive when Blastocystis was identified after Sanger and next-generation sequencing.
bSubtype information obtained from all positive samples via next-generation amplicon sequencing.
Molecular characterisation of Blastocystis
Among the 36 confirmed Blastocystis samples from wild boars in Spain, 31 were identified as ST5 by Sanger sequencing. Seventeen of them, plus five dubious samples (i.e., samples for which no Sanger sequencing data were obtained due to insufficient or poor-quality DNA), were subsequently analysed by NGS. Two Blastocystis subtypes (ST5 and ST15) were found by NGS among the 22 Blastocystis-positive samples analysed (Table 2 and Additional file 2). ST5 was the most prevalent Blastocystis ST identified (100%, 22/22) in this wild boar subpopulation, whereas ST15 was found in a single (4.5%, 1/22) isolate as a mixed coloniser with ST5 (Additional file 2).
Among the 34 confirmed Blastocystis samples from wild boars from Portugal, 24 were identified by Sanger sequencing as ST5. All of them, plus ten dubious samples for which no Sanger sequencing data were obtained, were subsequently analysed by NGS. Seven STs (ST5, ST10a, ST13, ST14, ST15, ST24b, and ST43) were identified among the 34 Blastocystis-positive samples (Table 3 and Additional file 2). Similar to the wild boar population from Spain, ST5 was the most prevalent ST identified in this subpopulation (100%, 34/34), followed by ST43 (5.9%, 2/34). The remaining STs identified (ST10a, ST13, ST15, ST15, and ST24b) were only rarely found (2.9% each, 1/34) and were always a mixed colonisation with ST5 (Table 3 and Additional file 2).
Blastocystis intra‑subtype diversity by NGS
Intra-subtype diversity was observed in only two STs, ST5 and ST15, the latter found in both Spain and Portugal, infecting one specimen each. No intra-subtype variability was detected within ST10a, ST13, ST14, ST24b, or ST43, where a single genetic variant was identified (Table 4). ST5 had the highest intra-subtype diversity, with eight unique genetic variants among the 56 Blastocystis-positive samples belonging to this ST. Four of them represented genetic variants shared between the Spanish and Portuguese populations. The remaining four were exclusively found circulating within the Spanish or Portuguese subpopulations (two each; Table 4 and Additional file 2).
Table 4.
Diversity of Blastocystis subtypes and unique genetic variants observed using next-generation amplicon sequencing (NGS) among Blastocystis-positive wild boars from Spain (n = 22) and Portugal (n = 34).
| Subtype | Subgroup | Samples (n) | Unique genetic variants (n) | Frequency of positive simples (%) | GenBank accession number(s)a |
|---|---|---|---|---|---|
| ST5 | 56 | 8 | 100 | OR730909(P)/OR730910(S) | |
| OR730911(P)/OR730912(S) | |||||
| OR730916(P)/OR730917(S) | |||||
| OR730918(P)/OR730919(S) | |||||
| OR730933(S) | |||||
| OR730938(S) | |||||
| OR730943(S) | |||||
| OR730947(P) | |||||
| ST10 | ST10a | 1 | 1 | 1.8 | OR730914(P) |
| ST13 | 1 | 1 | 1.8 | OR730913(P) | |
| ST14 | 1 | 1 | 1.8 | OR730946(P) | |
| ST15 | 2 | 2 | 3.6 | OR730944(S) | |
| OR730945(P) | |||||
| ST24 | ST24b | 1 | 1 | 1.8 | OR730915(P) |
| ST43 | 2 | 1 | 3.6 | OR730924(P) |
Subtypes previously reported in humans (regardless of their true zoonotic potential) are in bold.
a For unique variants identified in Spain and Portugal, two sequences were submitted to GenBank. The country in which the sequences were identified is denoted in parentheses by the GenBank accession number. P and S denote Portugal and Spain, respectively.
Mixed ST colonisations discriminated by NGS
Among the 56 positive samples sequenced in the present study via NGS, only 8 (14.3%) contained a co-colonisation encompassing ST5 with another Blastocystis ST. Mixed colonisations in wild boars from Spain were found in only one sample (4.5%, 1/22; Table 2 and Additional file 2), whereas they appeared to be more common in wild boars from Portugal (20.6%, 7/34; Table 3 and Additional file 2). Blastocystis colonisation by a single ST always involved ST5 regardless of the origin of the sampled animal (Additional file 2). The only wild boar from Spain harbouring a Blastocystis mixed ST colonisation presented ST5 and ST15. The seven Portuguese wild boars harbouring Blastocystis mixed ST colonisations presented up to seven distinct STs (ST5, ST10a, ST13, ST14, ST15, ST24b, and ST43) in six combinations (Table 3 and Additional file 2). However, in both countries, mixed colonisations encompassing two subtypes primarily carry ST5 (99.6–99.8%), whereas the remaining six STs were detected at residual (0.1–0.4%) carriage rates (Table 5).
Table 5.
Prevalence of Blastocystis subtype/subgroups and the means and ranges of subtype/subgroups detected in wild boars from Spain (SP) (n = 22) and Portugal (PT) (n = 34) using next-generation amplicon sequencing (NGS) in the present study.
| Subtype | Subtype prevalence (%) | Subtype reads (mean, %) | Subtype reads (range, %) | |||
|---|---|---|---|---|---|---|
| SP wild boar | PT wild boar | SP wild boar | PT wild boar | SP wild boar | PT wild boar | |
| ST5 | 100 | 100 | 100 | 100 | 99.6–100 | 99.8–100 |
| ST10a | 0 | 2.9 | – | 0.2 | – | 0.2 |
| ST13 | 0 | 2.9 | – | 0.1 | – | 0.1 |
| ST14 | 0 | 2.9 | – | 0.1 | – | 0.1 |
| ST15 | 4.5 | 2.9 | 0.4 | 0.1 | 0.4 | 0.1 |
| ST24b | 0 | 2.9 | – | 0.1 | – | 0.1 |
| ST43 | 0 | 5.9 | – | 0.2 | – | 0.1–0.2 |
Subtypes previously reported in humans (regardless of their true zoonotic potential) are in bold.
Discussion
This survey represents the largest attempt to assess the occurrence, molecular diversity, and zoonotic potential of Blastocystis subtypes in wild boars conducted in the Iberian Peninsula to date. Our study has several strengths, including a large sample size, broad geographic coverage, the use of highly sensitive molecular methods for detecting and discriminating Blastocystis genetic variants, and the assessment of the presence of mixed STs within a sample. The survey is also timely because information on the contribution of wild boar to Blastocystis epidemiology is scarce [21, 65] (Table 1). This ubiquitous protist has been detected in a wide range of domestic and wild animals, suggesting the potential for zoonotic transmission events in both directions (animal → human and human → animal) [66–71]. In Europe, prevalence rates in wild boars have been reported, ranging from 1–62% in free-living animals and 50–80% in captive animals. Globally, most of the Blastocystis cases documented in wild boars reported ST5 (79.7%, 184/231) (Table 1). ST5 is also the most widely reported ST in surveys conducted domestically [21], suggesting that this subtype is particularly well adapted to colonise members of the Suidae family. Our data revealed an overall Blastocystis colonisation rate of 15.3% in wild boars, with higher rates in wild boars from Portugal (34.3%) than in their counterparts from Spain (10.0%). These figures align with those estimated in a recent national study conducted in Portugal (29.0%, 42/144) [51]. However, a lower presence of Blastocystis (0.7%, 1/142) was detected in wild boar faeces in southern Spain [55].
In our study, NGS analyses confirmed the occurrence of seven distinct Blastocystis STs, including subgroup variants of ST10 and ST24 (ST5, ST10a, ST13, ST14, ST15, ST24b, and ST43), which circulate within the surveyed wild boar populations, with greater variability (in terms of genetic diversity and mixed STs colonisation rates) in wild boars from Portugal than in those from Spain. In addition to ST5, the remaining subtypes were missed by Sanger sequencing. While in Spain, only 4.5% (1/22) of the Blastocystis-positive wild boars identified by NGS harboured mixed colonisations, a much higher co-colonisation rate (20.6%, 7/34) was observed in their Portuguese counterparts. The reason for the higher prevalence and genetic variability rates observed in Portugal is unclear. Cross-species transmission involving other wildlife species (e.g., cervids) does not seem to be a plausible explanation, as no differences in the distribution of free-living species and management practices of natural protected/classified areas exist between the surveyed Spanish and Portuguese regions. However, free-roaming livestock herds can potentially act as local sources of Blastocystis in areas where sylvatic and domestic transmission cycles overlap. Indeed, in a parallel study targeting the same areas sampled in the present study, Blastocystis prevalence rates ranging from 56–80% were reported among cattle, sheep, and goats, and 22 distinct Blastocystis STs (including the ST10, ST24, and ST42 subgroups) were identified: ST1–ST3, ST5–ST7, ST10/b, ST13, ST14, ST21, ST23, ST24a/b/c, ST25, ST26, ST30, ST42a/b, ST43, and ST44 [42]. Similarly, cattle from Spain have been demonstrated to harbour up to 10 Blastocystis subtypes, including ST1, ST3, ST5, ST10, ST14, ST21, ST23, ST24, ST25, and ST26 using also NGS [41]. Taken together, these data might indicate that the presence (at low or very low rates) of Blastocystis STs other than ST5 in Iberian wild boars is the direct consequence of sporadic spillover events from livestock (primarily cattle) sharing habitats, most likely through environmental faecal contamination of water or grass fields. In fact, in addition to ST15 (also reported in Spanish wild boars), all Blastocystis STs identified in Portuguese wild boars were previously reported in livestock species from Portugal [42]. Cross-species transmission at the domestic‒wildlife interface has been previously demonstrated for other pathogens, such as Coxiella burnetii [72]. Additionally, supplemental feeding is a common practice in hunting states and game reserves in Mediterranean habitats and is usually related to the maintenance of artificial high population densities. This practice is known for increasing disease transmission risk in wildlife due to aggregation behaviours. However, it can also be used as a wildlife disease management option by delivering vaccines or anti-parasitic agents throughout the feed [73], which could explain the low Blastocystis prevalence and genetic diversity found in Spanish wild boars.
Our results revealed that wild boars in the Iberian Peninsula are suitable reservoirs for seven distinct Blastocystis STs (ST5, ST10a, ST13, ST14, ST15, ST24b, and ST43), of which ST5, ST10, and ST14 are potentially zoonotic. ST5 is the most prevalent ST reported in wild boar and domestic pigs worldwide, suggesting that swine are its natural host. Thus, ST5 has been detected in all but two of the studies that conducted Blastocystis subtyping in wild boars (Table 1). ST5 in wild boar has been documented in Brazil, Italy, Poland, Portugal, South Korea, Spain, and the United Kingdom (Table 1). Subtypes other than ST5 have also been detected in this host, including ST15 in wild boar faecal samples from Italy and Slovakia, as well as potentially zoonotic STs, including ST1, ST3, ST4, ST8, and ST10 [65] (Table 1). The presence of genetically diverse subtypes, representing differences in parasite-host preference, zoonotic potential, pathogenesis, and probably clinical manifestations, is another important issue associated with Blastocystis carriage. Human cases are primarily due to infection/colonisation by ST1–ST4; however, at least 12 additional STs (ST5–ST10, ST12, ST14, ST16, ST23, ST35, and ST41) have also been reported in human samples with varying frequencies [34–39]. From the “One Health” perspective, which links human, animal, and environmental health, a threat to any of the components of this triad can substantially impact the others [74]. Consequently, the probable presence of potential zoonotic Blastocystis STs in wild boars can influence humans and other animal species that share the same habitat.
This study had potential limitations that may have biased, at least partially, the results obtained. First, its retroactive nature required that some of the analysed faecal samples be stored at −20 °C for up to seven years before DNA extraction and molecular testing. Long-term storage may have altered the quantity/quality of parasite DNA, compromising the performance of the PCRs used for diagnostic and genotyping purposes. Second, owing to the legal hunting periods, our opportunistic sampling strategy limited our ability to capture potential seasonal variations in Blastocystis occurrence in wild boars. Third, the conventional PCR used for screening purposes lacks inhibition control. It is possible that an unknown number of our allegedly Blastocystis-negative samples indeed inhibited the PCR. Fourth, even though the sampling carried out in Spain was conducted nationwide, in Portugal, it was carried out only in the northeast and central areas of the country, taking advantage of ongoing projects, meaning that the results may not reflect the whole Portuguese scenario. Clearly, more research with a proper design should be conducted to disentangle how environmental, host, and management factors can modulate the risk of exposure of wild boar to Blastocystis.
This is the largest molecular epidemiological study investigating the presence and genetic diversity of Blastocystis in wild boars conducted in the Iberian Peninsula to date. Overall, the presence of Blastocystis was relatively low (10%) in wild boars from Spain and was caused mainly by swine-adapted ST5. The opposite scenario was found in Portugal, with a much higher prevalence (34.3%) and genetic diversity (up to 7 STs), indicative of possible cross-species transmission or contamination from free-ranging livestock animals that share habitats. Our results show that wild boars, which are most likely in contact with domestic ungulates and possibly other wild animals, are important reservoirs of Blastocystis in the Iberian Peninsula. However, spurious infections (e.g., those expected in highly anthropized environments such as agricultural and peri-urban areas) cannot be ruled out. In this sense, adopting regular monitoring programs, encompassing the sampling of both wild and domestic animals, with more extensive national coverage and sampling sites, involving hunting associations and other partners (universities, national labs) to increase sample collection and storage, may help us obtain a better picture of the Blastocystis epidemiological scenario in the Iberian Peninsula, as well as a wide array of other protists and zoonoses, and its potential transmission risks for the human compartment.
Supplementary Information
Additional file 1. Summary of the sampling sites in Portugal according to bioregion with an emphasis on environmental, wildlife and flora features, adapted from PNVSFS (2020) and Muñoz et al. [58]. The numbers of wild boar faecal samples collected at each location are indicated.
Additional file 2. Full dataset showing the epidemiological data used in the analyses conducted in this study, as well as the diagnostic and molecular results obtained.
Acknowledgements
This publication is based upon work from COST Action Blastocystis under One Health, CA21105, supported by COST (European Cooperation in Science and Technology). Sampling in the Basque Country was conducted by members of the Association for the Defence of the Game Natural Heritage of the Basque Country (ARTIO). We thank the Dirección General del Medio Natural y Planificación Rural del Principado de Asturias (Oviedo, Spain). We thank Aleksey Molokin and Nadja S. George for their technical assistance. Wildlife Ecology & Health Group (WE&H): The following investigators (in alphabetic order) participated in the Wildlife Ecology & Health Group (WE&H): Carles Conejero, Carlos González-Crespo, Cristina Garrido, Diana Gassó, Emmanuel Serrano, Gregorio Mentaberre, Irene Torres-Blas, Josep Estruch, Josep Pastor, Jorge Ramón López-Olvera, María Escobar-González, Marta Valldeperes, Montse Mesalles, Rafaela Cuenca, Roser Velarde, Santiago Lavín.
Authors' contributions
AMF, MAH, ARJ, JV, MCA, DFL, PM, JAA, AB, GAC, RCB, JC, DH, JF, JDP, DGB and ES (on behalf of the WE&H group) collected the samples. AD, AMF, PCK, BB and JGM carried out the laboratory experiments. DGB, MS and DC designed and supervised the experiments. PCK, AMF, DGB, MS and DC wrote the original draft. RTT, CF, AM, ARJ, AB, RCB, ES, DGB, MS and DC writing—review and editing. The final version was read and approved by all the authors.
Funding
This study was partially funded by the Health Institute Carlos III (ISCIII), Spanish Ministry of Economy and Competitiveness, under project PI19CIII/00029 and the USDA-ARS Project No: 8042-32000-100-00-D. A.M.F., D.H. and J.F. were supported by PhD grants from the Fundação para a Ciência e Tecnologia (SFRH/BD/144582/2019, SFRH/BD/144437/2019, PD/BD/150645/2020, respectively), co-financed by the European Social Fund POPH-QREN programme. R.T.T. and J.C. were supported by Research Contracts from the Fundação para a Ciência e a Tecnologia (2021.00690. CEECIND and CEECIND/01428/2018, respectively). A.R.-J. is the recipient of a Miguel Servet Research Contract funded by the Spanish Ministry of Science, Innovation and Universities (CP18/00111). D.G.B. is the recipient of a Sara Borrell Research Contract (CD19CIII/00011) funded by the Spanish Ministry of Science, Innovation and Universities. A.D. is the recipient of a PFIS contract (FI20CIII/00002) funded by the Spanish Ministry of Science, Innovation and Universities.
Availability of data and materials
The data that support the findings of this study are available within the main body of the manuscript and its supplementary material.
Declarations
Ethics approval and consent to participate
Sampled animals were legally hunted under Spanish, Portuguese and EU (RD 8/2003, RD 173/99, 38/2020; RD 53/2013) legislation. All the hunters had hunting licences, and no animal was hunted for the project’s sake but for annual hunting activities following the abovementioned legislation. Professional personnel collected faecal samples from hunter-harvested wild boars during the regular hunting season.
Competing interests
The authors declare that they have no competing interests.
Footnotes
The original version of this article was revised: the authors updated the Acknowledgements section.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pamela C. Köster and Ana M. Figueiredo contributed equally to this article.
Emmanuel Serrano—On behalf of the WE&H group. The members of the group are listed at the Acknowledgements section.
Change history
11/14/2024
A Correction to this paper has been published: 10.1186/s13567-024-01408-5
Contributor Information
David González-Barrio, Email: david.gonzalezb@isciii.es.
Monica Santin, Email: monica.santin-duran@usda.gov.
References
- 1.Choi SK, Kim KS, Ranyuk M, Babaev E, Voloshina I, Bayarlkhagva D, Chong JR, Ishiguro N, Yu L, Min MS, Lee H, Markov N (2020) Asia-wide phylogeography of wild boar (Sus scrofa) based on mitochondrial DNA and Y-chromosome: revising the migration routes of wild boar in Asia. PLoS One 15:e0238049. 10.1371/journal.pone.0238049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massei G, Kindberg J, Licoppe A, Gačić D, Šprem N, Kamler J, Baubet E, Hohmann U, Monaco A, Ozoliņš J, Cellina S, Podgórski T, Fonseca C, Markov N, Pokorny B, Rosell C, Náhlik A (2015) Wild boar populations up, numbers of hunters down? a review of trends and implications for Europe. Pest Manag Sci 71:492–500. 10.1002/ps.3965 [DOI] [PubMed] [Google Scholar]
- 3.Valente AM, Acevedo P, Figueiredo AM, Fonseca C, Torres RT (2020) Overabundant wild ungulate populations in Europe: management with consideration of socio-ecological consequences. Mamm Rev 50:353–366. 10.1111/mam.12202 [Google Scholar]
- 4.Casas-Díaz E, Closa-Sebastià F, Peris A, Serrano E (2013) Recorded dispersal of wild boar (Sus scrofa) in Northeast Spain: implications for disease-monitoring programs. Wildl Biol Pract 9:19–26. 10.2461/wbp.2013.ibeun.3 [Google Scholar]
- 5.Castillo-Contreras R, Carvalho J, Serrano E, Mentaberre G, Fernández-Aguilar X, Colom A, González-Crespo C, Lavín S, López-Olvera JR (2018) Urban wild boars prefer fragmented areas with food resources near natural corridors. Sci Total Environ 615:282–288. 10.1016/j.scitotenv.2017.09.277 [DOI] [PubMed] [Google Scholar]
- 6.Apollonio M, Andersen R, Putman R (2010) European ungulates and their management in the 21st century. Cambridge University Press [Google Scholar]
- 7.Linnell JD, Cretois B, Nilsen EB, Rolandsen CM, Solberg EJ, Veiberg V, Kaczensky P, Van Moorter B, Panzacchi M, Rauset GR, Kaltenborn B (2020) The challenges and opportunities of coexisting with wild ungulates in the human-dominated landscapes of Europe’s anthropocene. Biol Conserv 244:108500. 10.1016/j.biocon.2020.108500 [Google Scholar]
- 8.Lagos L, Picos J, Valero E (2012) Temporal pattern of wild ungulate-related traffic accidents in northwest Spain. Eur J Wildl Res 58:661–668. 10.1007/s10344-012-0614-6 [Google Scholar]
- 9.Torres RT, Linck P, Pinto N, Ares-Pereira G, Barroqueiro C, Fonseca C, Carvalho J (2023) Landscape and population drivers of ungulate-vehicle collisions in Portugal. Appl Geogr 151:102859. 10.1016/j.apgeog.2022.102859 [Google Scholar]
- 10.Herrero J, García-Serrano A, Couto S, Ortuño VM, García-González R (2006) Diet of wild boar Sus scrofa L. and crop damage in an intensive agroecosystem. Eur J Wildl Res 52:245–250. 10.1007/s10344-006-0045-3 [Google Scholar]
- 11.Carpio AJ, Guerrero-Casado J, Ruiz-Aizpurua L, Vicente J, Tortosa FS (2014) The high abundance of wild ungulates in a Mediterranean region: Is this compatible with the European rabbit? Wildl Biol 20:161–166. 10.2981/wlb.13113 [Google Scholar]
- 12.Carpio AJ, Guerrero-Casado J, Tortosa FS, Vicente J (2014) Predation of simulated red-legged partridge nests in big game estates from South Central Spain. Eur J Wildl Res 60:391–394. 10.1007/s10344-013-0786-8 [Google Scholar]
- 13.Meng XJ, Lindsay DS, Sriranganathan N (2009) Wild boars as sources for infectious diseases in livestock and humans. Philos Trans R Soc Lond B Biol Sci 364:2697–2707. 10.1098/rstb.2009.0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz-Fons F (2017) A review of the current status of relevant zoonotic pathogens in wild swine (Sus scrofa) populations: changes modulating the risk of transmission to humans. Transbound Emerg Dis 64:68–88. 10.1111/tbed.12369 [DOI] [PubMed] [Google Scholar]
- 15.Abrantes AC, Vieira-Pinto M (2023) 15 years overview of European zoonotic surveys in wild boar and red deer: a systematic review. One Health 16:100519. 10.1016/j.onehlt.2023.100519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gortázar C, Torres MJ, Vicente J, Acevedo P, Reglero M, De la Fuente J, Negro JJ, Aznar-Martín J (2008) Bovine tuberculosis in Doñana Biosphere Reserve: the role of wild ungulates as disease reservoirs in the last Iberian lynx strongholds. PLoS One 3:e2776. 10.1371/journal.pone.0002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barroso P, Serrano E, Carpio AJ, AcevedoP VJ, Gortázar C (2023) Low impact of tuberculosis severity on wild boar body condition. Res Vet Sci 155:161–167. 10.1016/j.rvsc.2023.01.014 [DOI] [PubMed] [Google Scholar]
- 18.Postel A, Austermann-Busch S, Petrov A, Moennig V, Becher P (2018) Epidemiology, diagnosis and control of classical swine fever: recent developments and future challenges. Transbound Emerg Dis 65:248–261. 10.1111/tbed.12676 [DOI] [PubMed] [Google Scholar]
- 19.Ruiz-Fons F, Vidal D, Höfle U, Vicente J, Gortázar C (2007) Aujeszky’s disease virus infection patterns in European wild boar. Vet Microbiol 120:241–250. 10.1016/j.vetmic.2006.11.003 [DOI] [PubMed] [Google Scholar]
- 20.Jota Baptista C, Seixas F, Gonzalo-Orden JM, Oliveira PA (2023) Wild boar (Sus scrofa) as a potential reservoir of infectious agents in Portugal: a review of two decades (2001–2021). Eur J Wildl Res 69:101. 10.1007/s10344-023-01732-9 [Google Scholar]
- 21.Hublin JSY, Maloney JG, Santin M (2021) Blastocystis in domesticated and wild mammals and birds. Res Vet Sci 135:260–282. 10.1016/j.rvsc.2020.09.031 [DOI] [PubMed] [Google Scholar]
- 22.Stensvold CR, Clark CG (2020) Pre-empting Pandora’s box: Blastocystis subtypes revisited. Trends Parasitol 36:229–232. 10.1016/j.pt.2019.12.009 [DOI] [PubMed] [Google Scholar]
- 23.Reh L, Muadica AS, Köster PC, Balasegaram S, Verlander NQ, Chércoles ER, Carmena D (2019) Substantial prevalence of enteroparasites Cryptosporidium spp., Giardiaduodenalis and Blastocystis sp. in asymptomatic schoolchildren in Madrid, Spain, November 2017 to June 2018. Euro Surveill 24:1900241. 10.2807/1560-7917.ES.2019.24.43.1900241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scanlan PD, Stensvold CR, Rajilić-Stojanović M, Heilig HG, De Vos WM, O’Toole PW, Cotter PD (2014) The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol Ecol 90:326–330. 10.1111/1574-6941.12396 [DOI] [PubMed] [Google Scholar]
- 25.Audebert C, Even G, Cian A, Loywick A, Merlin S, Viscogliosi E, Chabé M, Blastocystis Investigation Group (2016) Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci Rep 6:25255. 10.1038/srep25255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beghini F, Pasolli E, Truong TD, Putignani L, Cacciò SM, Segata N (2017) Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J 11:2848–2863. 10.1038/ismej.2017.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tito RY, Chaffron S, Caenepeel C, Lima-Mendez G, Wang J, Vieira-Silva S, Falony G, Hildebrand F, Darzi Y, Rymenans L, Verspecht C, Bork P, Vermeire S, Joossens M, Raes J (2019) Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut 68:1180–1189. 10.1136/gutjnl-2018-316106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maloney JG, Molokin A, Seguí R, Maravilla P, Martínez-Hernández F, Villalobos G, Tsaousis AD, Gentekaki E, Muñoz-Antolí C, Klisiowicz DR, Oishi CY, Toledo R, Esteban JG, Köster PC, de Lucio A, Dashti A, Bailo B, Calero-Bernal R, González-Barrio D, Carmena D, Santín M (2022) Identification and molecular characterization of four new Blastocystis subtypes designated ST35-ST38. Microorganisms 11:46. 10.3390/microorganisms11010046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santin M, Molokin A, Maloney JG (2023) A longitudinal study of Blastocystis in dairy calves from birth through 24 months demonstrates dynamic shifts in infection rates and subtype prevalence and diversity by age. Parasit Vectors 16:177. 10.1186/s13071-023-05795-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koehler AV, Herath HMPD, Hall RS, Wilcox S, Gasser RB (2023) Marked genetic diversity within Blastocystis in Australian wildlife revealed using a next generation sequencing-phylogenetic approach. Int J Parasitol Parasites Wildl 23:100902. 10.1016/j.ijppaw.2023.100902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Šejnohová A, Koutenská M, Jirků M, Brožová K, Pavlíčková Z, Kadlecová O, Cinek O, Maloney JG, Santín M, Petrželková KJ, Jirků K (2024) A cross-sectional survey of Blastocystis sp. and Dientamoebafragilis in non-human primates and their caregivers in Czech zoos. One Health 19:100862. 10.1016/j.onehlt.2024.100862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santin M, Figueiredo A, Molokin A, George N, Köster PC, Dashti A, González-Barrio D, Carmena D, Maloney JG (2024) Division of Blastocystis ST10 into three new subtypes: ST42-ST44. J Eukaryot Microbiol 1:e12998 [DOI] [PubMed] [Google Scholar]
- 33.Stensvold RC, Berg RPKD, Maloney JG, Molokin A, Santin M (2023) Molecular characterization of Blastocystis and Entamoeba of muskoxen and sheep in Greenland. Int J Parasitol 53:673–685. 10.1016/j.ijpara.2023.05.005 [DOI] [PubMed] [Google Scholar]
- 34.Ramírez JD, Sánchez A, Hernández C, Flórez C, Bernal MC, Giraldo JC, Reyes P, López MC, García L, Cooper PJ, Vicuña Y, Mongi F, Casero RD (2016) Geographic distribution of human Blastocystis subtypes in South America. Infect Genet Evol 41:32–35. 10.1016/j.meegid.2016.03.017 [DOI] [PubMed] [Google Scholar]
- 35.Khaled S, Gantois N, Ly AT, Senghor S, Even G, Dautel E, Dejager R, Sawant M, Baydoun M, Benamrouz-Vanneste S, Chabé M, Ndiaye S, Schacht AM, Certad G, Riveau G, Viscogliosi E (2020) Prevalence and subtype distribution of Blastocystis sp. in Senegalese school children. Microorganisms 8:1408. 10.3390/microorganisms8091408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jinatham V, Maxamhud S, Popluechai S, Tsaousis AD, Gentekaki E (2021) Blastocystis One Health approach in a rural community of northern Thailand: prevalence, subtypes and novel transmission routes. Front Microbiol 12:746340. 10.3389/fmicb.2021.746340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khaled S, Gantois N, Ayoubi A, Even G, Sawant M, El Houmayraa J, Nabot M, Benamrouz-Vanneste S, Chabé M, Certad G, El Safadi D, Dabboussi F, Hamze M, Viscogliosi E (2021) Blastocystis sp. prevalence and subtypes distribution amongst Syrian refugee communities living in north Lebanon. Microorganisms 9:184. 10.3390/microorganisms9010184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osorio-Pulgarin MI, Higuera A, Beltran-Álzate JC, Sánchez-Jiménez M, Ramírez JD (2021) Epidemiological and molecular characterization of Blastocystis infection in children attending daycare centers in Medellín. Colombia Biology 10:669. 10.3390/biology10070669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernández-Castro C, Maloney JG, Agudelo-López SP, Toro-Londoño MA, Botero-Garcés JH, Orozco MC, Quintero-Quinchia YC, Correa-Cote JC, Múnera-Duque A, Ricaurte-Ciro JC, Londoño-Álvarez LI, Escobar RM, Köster PC, Sánchez S, Carmena D, Santín M (2023) Identification and validation of novel Blastocystis subtype ST41 in a Colombian patient undergoing colorectal cancer screening. J Eukaryot Microbiol 70:e12978 [DOI] [PubMed] [Google Scholar]
- 40.Maloney JG, Molokin A, Santin M (2019) Next generation amplicon sequencing improves detection of Blastocystis mixed subtype infections. Infect Genet Evol 73:119–125. 10.1016/j.meegid.2019.04.013 [DOI] [PubMed] [Google Scholar]
- 41.Abarca N, Santín M, Ortega S, Maloney JG, George NS, Molokin A, Cardona GA, Dashti A, Köster PC, Bailo B, Hernández-de-Mingo M, Muadica AS, Calero-Bernal R, Carmena D, González-Barrio D (2021) Molecular detection and characterization of Blastocystis sp. and Enterocytozoonbieneusi in cattle in northern Spain. Vet Sci 8:191. 10.3390/vetsci8090191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Figueiredo AM, Santín M, Köster PC, Dashti A, Maloney JG, Torres RT, Fonseca C, Mysterud A, Carvalho J, Hipólito D, Rossa M, Palmeira JD, González-Barrio D, Calero-Bernal R, Carmena D (2024) Molecular detection and characterization of Blastocystis in herbivore livestock species in Portugal. Vet Parasitol 327:110147. 10.1016/j.vetpar.2024.110147 [DOI] [PubMed]
- 43.Mundim MJS, Mundim AV, Santos ALQ, Cabral DD, Faria ESM, Moraes FM (2004) Helminths and protozoa in wild boars (Sus scrofa scrofa) feces raised in captivity. Arq Bras Med Vet Zootec 56:792–795. 10.1590/S0102-09352004000600015 [Google Scholar]
- 44.Valença-Barbosa C, de Bomfim TCB, Teixeira BR, Gentile R, Neto SFDC, Magalhães BSN, Balthazar DA, da Silva FA, Biot R, d’Avila Levy CM, Santos HLC (2019) Molecular epidemiology of Blastocystis isolated from animals in the state of Rio de Janeiro, Brazil. PLoS One 14:e0210740. 10.1371/journal.pone.0210740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, Meng W, Zhou Z, Deng L, Shi X, Chai Y, Liu H, Cheng Y, Zhong Z, Fu H, Shen L, Zhang K, He T, Peng G (2021) Genetic characterization and zoonotic potential of Blastocystis from wild animals in Sichuan Wolong National Natural Reserve, Southwest China. Parasite 28:73. 10.1051/parasite/2021071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solaymani-Mohammadi S, Rezaian M, Hooshyar H, Mowlavi GR, Babaei Z, Anwar MA (2004) Intestinal protozoa in wild boars (Sus scrofa) in western Iran. J Wildl Dis 40:801–803. 10.7589/0090-3558-40.4.801 [DOI] [PubMed] [Google Scholar]
- 47.Yaghoobi K, Sarkari B, Mansouri M, Motazedian MH (2016) Zoonotic intestinal protozoan of the wild boars, Sus scrofa, in Persian Gulf’s coastal area (Bushehr province), Southwestern Iran. Vet World 9:1047–1050. 10.14202/vetworld.2016.1047-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russini V, Di Filippo MM, Fanelli R, Polidori M, Berrilli F, Di Cave D, Novelletto A, Calderini P (2020) Characterization of prevalence and genetic subtypes of Blastocystis sp. in wild and domestic Suidae of central Italy aided by amplicon NGS. Vet Parasitol Reg Stud Reports 22:100472. 10.1016/j.vprsr.2020.100472 [DOI] [PubMed] [Google Scholar]
- 49.Kaczmarek A, Sobociński W, Wesołowska M, Gołąb E, Kołodziej-Sobocińska M, Sałamatin R (2021) Blastocystis occurrence and subtype diversity in wild European terrestrial mammals—the case of Białowieża Primeval Forest (NE Poland). Int J Parasitol Parasites Wildl 16:120–125. 10.1016/j.ijppaw.2021.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudzińska M, Kowalewska B, Waleron M, Kalicki M, Sikorska K, Szostakowska B (2021) Molecular characterization of Blastocystis from animals and their caregivers at the Gdańsk Zoo (Poland) and the assessment of zoonotic transmission. Biology 10:984. 10.3390/biology10100984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santos-Silva S, Moraes DFDSD, López-López P, Palmeira JD, Torres RT, São José Nascimento M, Dashti A, Carmena D, Rivero-Juarez A, Mesquita JR (2023) Survey of zoonotic diarrheagenic protist and hepatitis E virus in wild boar (Sus scrofa) of Portugal. Animals 13:256. 10.3390/ani13020256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danišová O, Valenčáková A, Kandráčová P, Tomko M, Sučik M (2022) First report of Blastocystis spp. subtypes in Zoo animals in Slovakia, Central Europe. Ann Agric Environ Med 29:149–151. 10.26444/aaem/145826 [DOI] [PubMed] [Google Scholar]
- 53.Valenčáková A, Sučik M, Danišová O, Kandráčová P, Tomko M, Valocky I (2022) Detection of Blastocystis spp., Cryptosporidium spp. and Encephalitozoon spp. among wild animals from Eastern Slovakia. Acta Vet Hung. 10.1556/004.2022.00026 [DOI] [PubMed] [Google Scholar]
- 54.Lee H, Seo MG, Oem JK, Kim YS, Lee SY, Kim J, Jeong H, Jheong WH, Kim Y, Lee WJ, Kwon OD, Kwak D (2020) Molecular detection and subtyping of Blastocystis detected in wild boars (Sus scrofa) in South Korea. J Wildl Dis 56:662–666. 10.7589/2019-04-092 [DOI] [PubMed] [Google Scholar]
- 55.Rivero-Juárez A, Dashti A, Santín M, Köster PC, López-López P, Risalde MA, García-Bocanegra I, Gómez-Villamandos JC, Caballero-Gómez J, Frías M, Bailo B, Ortega S, Muadica AS, Calero-Bernal R, González-Barrio D, Rivero A, Briz V, Carmena D (2022) Diarrhoea-causing enteric protist species in intensively and extensively raised pigs (Sus scrofa domesticus) in southern Spain. Part II: association with hepatitis E virus susceptibility. Transbound Emerg Dis 69:e1172–e1178. 10.1111/tbed.14408 [DOI] [PubMed] [Google Scholar]
- 56.Betts EL, Gentekaki E, Thomasz A, Breakell V, Carpenter AI, Tsaousis AD (2018) Genetic diversity of Blastocystis in non-primate animals. Parasitology 145:1228–1234. 10.1017/S0031182017002347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Betts EL, Gentekaki E, Tsaousis AD (2020) Exploring micro-eukaryotic diversity in the gut: Co-occurrence of Blastocystis subtypes and other protists in zoo animals. Front Microbiol 11:288. 10.3389/fmicb.2020.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muñoz PM, Boadella M, Arnal M, de Miguel MJ, Revilla M, Martínez D, Vicente J, Acevedo P, Oleaga A, Ruiz-Fons F, Marín CM, Prieto JM, de la Fuente J, Barral M, Barberán M, de Luco DF, Blasco JM, Gortázar C (2010) Spatial distribution and risk factors of Brucellosis in Iberian wild ungulates. BMC Infect Dis 10:46. 10.1186/1471-2334-10-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.PNVSFS, Plan Nacional de Vigilancia Sanitaria en Fauna Silvestre, MAPA, Ministerio de Agricultura, Pesca y Alimentación; 2020. https://www.mapa.gob.es/es/ganaderia/temas/sanidad-animal-higiene-ganadera/pvfs2020_tcm30-437517.pdf. Accessed 23 Aug 2024.
- 60.Direção Geral de Alimentação e Veterinária DGAV. https://www.dgav.pt
- 61.Natura; 2000. https://ec.europa.eu/environment/nature/natura2000/index_en.htm
- 62.Scicluna SM, Tawari B, Clark CG (2006) DNA barcoding of Blastocystis. Protist 157:77–85. 10.1016/j.protis.2005.12.001 [DOI] [PubMed] [Google Scholar]
- 63.Santín M, Gómez-Muñoz MT, Solano-Aguilar G, Fayer R (2011) Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol Res 109:205–212. 10.1007/s00436-010-2244-9 [DOI] [PubMed] [Google Scholar]
- 64.R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2022. https://www.R-project.org/.
- 65.Asghari A, Sadrebazzaz A, Shamsi L, Shams M (2021) Global prevalence, subtypes distribution, zoonotic potential, and associated risk factors of Blastocystis sp. in domestic pigs (Susdomesticus) and wild boars (Susscrofa): a systematic review and meta-analysis. Microb Pathog 160:105183. 10.1016/j.micpath.2021.105183 [DOI] [PubMed] [Google Scholar]
- 66.Parkar U, Traub RJ, Kumar S, Mungthin M, Vitali S, Leelayoova S, Morris K, Thompson RC (2007) Direct characterization of Blastocystis from faeces by PCR and evidence of zoonotic potential. Parasitology 134:359–367. 10.1017/S0031182006001582 [DOI] [PubMed] [Google Scholar]
- 67.Stensvold CR, Alfellani MA, Nørskov-Lauritsen S, Prip K, Victory EL, Maddox C, Nielsen HV, Clark CG (2009) Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int J Parasitol 39:473–479. 10.1016/j.ijpara.2008.07.006 [DOI] [PubMed] [Google Scholar]
- 68.Greige S, El Safadi D, Bécu N, Gantois N, Pereira B, Chabé M, Benamrouz-Vanneste S, Certad G, El Hage R, Chemaly M, Hamze M, Viscogliosi E (2018) Prevalence and subtype distribution of Blastocystis sp isolates from poultry in Lebanon and evidence of zoonotic potential. Parasit Vectors 11:389. 10.1186/s13071-018-2975-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li LH, Zhang XP, Lv S, Zhang L, Yoshikawa H, Wu Z, Steinmann P, Utzinger J, Tong XM, Chen SH, Zhou XN (2007) Cross-sectional surveys and subtype classification of human Blastocystis isolates from four epidemiological settings in China. Parasitol Res 102:83–90. 10.1007/s00436-007-0727-0 [DOI] [PubMed] [Google Scholar]
- 70.Wang W, Owen H, Traub RJ, Cuttell L, Inpankaew T, Bielefeldt-Ohmann H (2014) Molecular epidemiology of Blastocystis in pigs and their in-contact humans in Southeast Queensland, Australia, and Cambodia. Vet Parasitol 203:264–269. 10.1016/j.vetpar.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 71.Yan Y, Su S, Ye J, Lai X, Lai R, Liao H, Chen G, Zhang R, Hou Z, Luo X (2007) Blastocystis sp. subtype 5: a possibly zoonotic genotype. Parasitol Res 101:1527–1532. 10.1007/s00436-007-0672-y [DOI] [PubMed] [Google Scholar]
- 72.González-Barrio D, Carpio AJ, Sebastián-Pardo M, Peralbo-Moreno A, Ruiz-Fons F (2022) The relevance of the wild reservoir in zoonotic multi-host pathogens: The links between Iberian wild mammals and Coxiella burnetii. Transbound Emerg Dis 69:3868–3880. 10.1111/tbed.14758 [DOI] [PubMed] [Google Scholar]
- 73.Sorensen A, van Beest FM, Brook RK (2014) Impacts of wildlife baiting and supplemental feeding on infectious disease transmission risk: a synthesis of knowledge. Prev Vet Med 113:356–363. 10.1016/j.prevetmed.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 74.Shah SS, Khan A (2019) One health and parasites. In: Yasobant S, Saxena D (eds) Global applications of one health practice and care. IGI Global, pp 82–112 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Summary of the sampling sites in Portugal according to bioregion with an emphasis on environmental, wildlife and flora features, adapted from PNVSFS (2020) and Muñoz et al. [58]. The numbers of wild boar faecal samples collected at each location are indicated.
Additional file 2. Full dataset showing the epidemiological data used in the analyses conducted in this study, as well as the diagnostic and molecular results obtained.
Data Availability Statement
The data that support the findings of this study are available within the main body of the manuscript and its supplementary material.