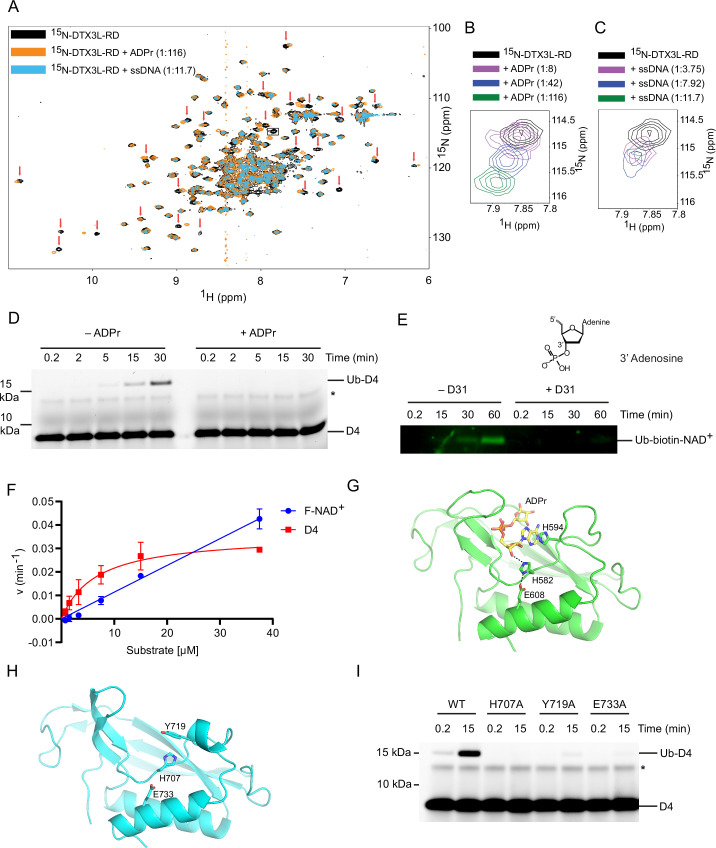
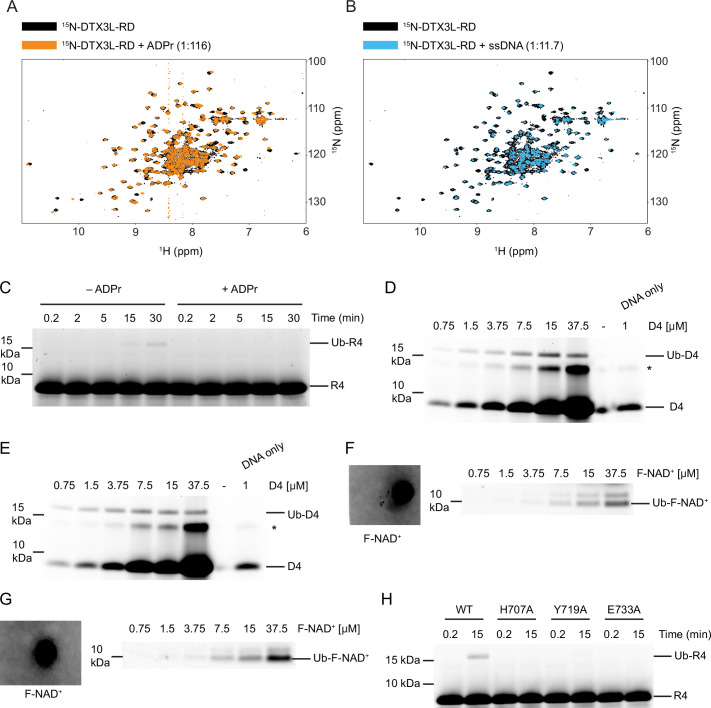
Figure 4. DTX3L DTC domain binds and facilitates ubiquitin (Ub)-DNA formation.
(A) 1H-15N heteronuclear single-quantum coherence (HSQC) spectra of 15N-DTX3L-RD (black), ADPr-15N-DTX3L-RD (orange), and single-stranded DNA (ssDNA) D30-15N-DTX3L-RD (blue). Red arrows indicate cross peaks that shift upon titrating with adenosine 5′-diphosphate (ADP)–ribose (ADPr) or ssDNA. (B) Close-up view of the cross peak indicated by the black box in (A) upon titration of specified molar ratios of ADPr with 15N-DTX3L-RD. (C) Close-up view of the cross peak indicated by the black arrow in (A) upon titration of specified molar ratios of ssDNA D30 with 15N-DTX3L-RD. (D) Fluorescently detected SDS-PAGE gel of in vitro ubiquitination of 6-FAM-labelled ssDNA D4 by DTX3L-RD in the presence of E1, UBE2D2, Ub, Mg2+-ATP and treated with excess ADPr. (E) Western blot of in vitro ubiquitination of biotin-NAD+ by DTX3L-RD in the presence of E1, UBE2D2, Ub, Mg2+-ATP and treated with excess ssDNA D31. (F) Kinetics of Ub-D4 and Ub-F-NAD+ formation catalysed by DTX3L-RD. Data from two independent experiments (n=2) were fitted with the Michaelis–Menten equation and kcat/Km value for D4 (5457 M–1 min–1) was calculated. kcat/Km value for F-NAD+ (1190 M–1 min–1) was estimated from the slope of the linear portion of the curve. (G) Structure of DTX2-DTC domain (green) bound to ADPr (yellow) (PDB: 6Y3J). The sidechains of H582, H594, and E608 are shown in sticks. Hydrogen bonds are indicated by dotted lines. (H) Structure of DTX3L-DTC domain (cyan; PDB: 3PG6). The sidechains of H707, Y719, and E733 are shown in sticks. (I) Fluorescently detected SDS-PAGE gel of in vitro ubiquitination of 6-FAM-labelled ssDNA D4 by full length DTX3L WT, H707A, Y719A, and E733A in the presence of E1, UBE2D2, Ub, Mg2+-ATP. Asterisks in (D) and (I) indicate contaminant band from ssDNA. Raw unedited and uncropped gel images of (D), (E) and (I) are shown in Figure 4—source data 1 and 2, respectively. Data points for (F) are shown in Figure 4—source data 3.