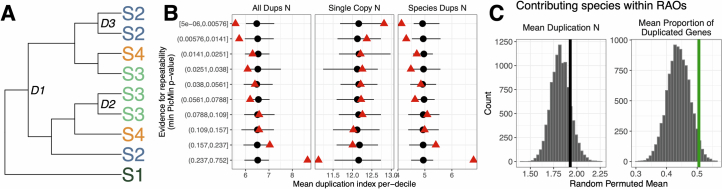
Extended Data Fig. 8. Associations between repeatability and gene duplication within orthogroups.
Panel A shows a simplified gene tree with 4 species (S1-S4), 3 duplications (D1-3), and 2 species-specific duplications (D2-3). Panel B shows depletion of duplications in orthogroups grouped according to their strongest evidence of repeatability. The mean duplication value is plotted for each repeatability decile. The black points and lines show a comparable analysis over 100 randomisations of all per gene ep-valuesWZA. The black point shows the mean duplication metric per decile and the lines show the maximum and minimum values across the 100 randomisations. Panel C shows duplication results exclusively looking at contributing species within RAOs, compared with species with low ep-valuesWZA (<0.1) within 10,000 draws of random orthogroup-climate sets. Lines show the observed mean relative to the distribution of random means, where green lines show permuted p-value < 0.05. The one-sided p-values for permutation tests = 0.153 (Mean Duplication N) and 0.035 (Mean Proportion of Duplicated Genes).