Fig. 4.
Molecular dynamics (MD) simulation of the interaction between anthricin and proteasome β-ring
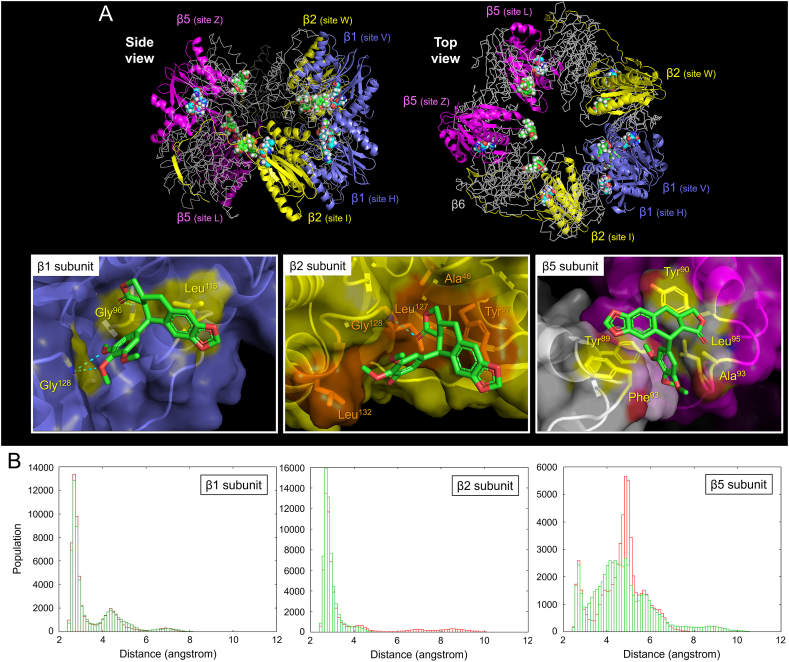
(A) MD simulations were performed based on the proteasome's crystal structure (Saccharomyces cerevisiae) complexed with K-7174, a proteasome inhibitor with similar structure to anthricin (PDB ID: 4EU2), by replacing K-7174 with anthricin. The representative complex structure after 200 ns simulation is shown. The β1, β2, and β5 subunits involved in enzyme activity are depicted in ribbons, whereas the other β-subunits are shown in gray lines. The three amino acid residues at the catalytic sites (Thr1, Asp17, and Lys33) of the β1, β2, and β5 subunits are shown as light blue spheres, and anthricin is depicted as green spheres. In the close-up view of the anthricin binding site, anthricin is shown as green sticks, whereas the interacting residues of the β1 subunit (Gly96, Gly128 and Leu115), the β2 subunit (Ala46, Tyr97, Leu127, Gly128 and Leu132), the β5 subunit (Tyr90, Ala93 and Leu95), and the β6 subunit (Tyr89 and Phe93) are shown as yellow or orange color sticks. (B) The three bar graphs plot the distances between the side chains of Thr1, Asp17, and Lys33 at the catalytic sites of the β1, β2, and β5 subunits during the 200 ns simulation. The red bars represent the apo form, whereas the green bars represent the proteasome bound with anthricin. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)