Fig. 2 |. Phylogenetic tree and assembly statistics of genomes assembled using the VGP–Galaxy assembly pipeline.
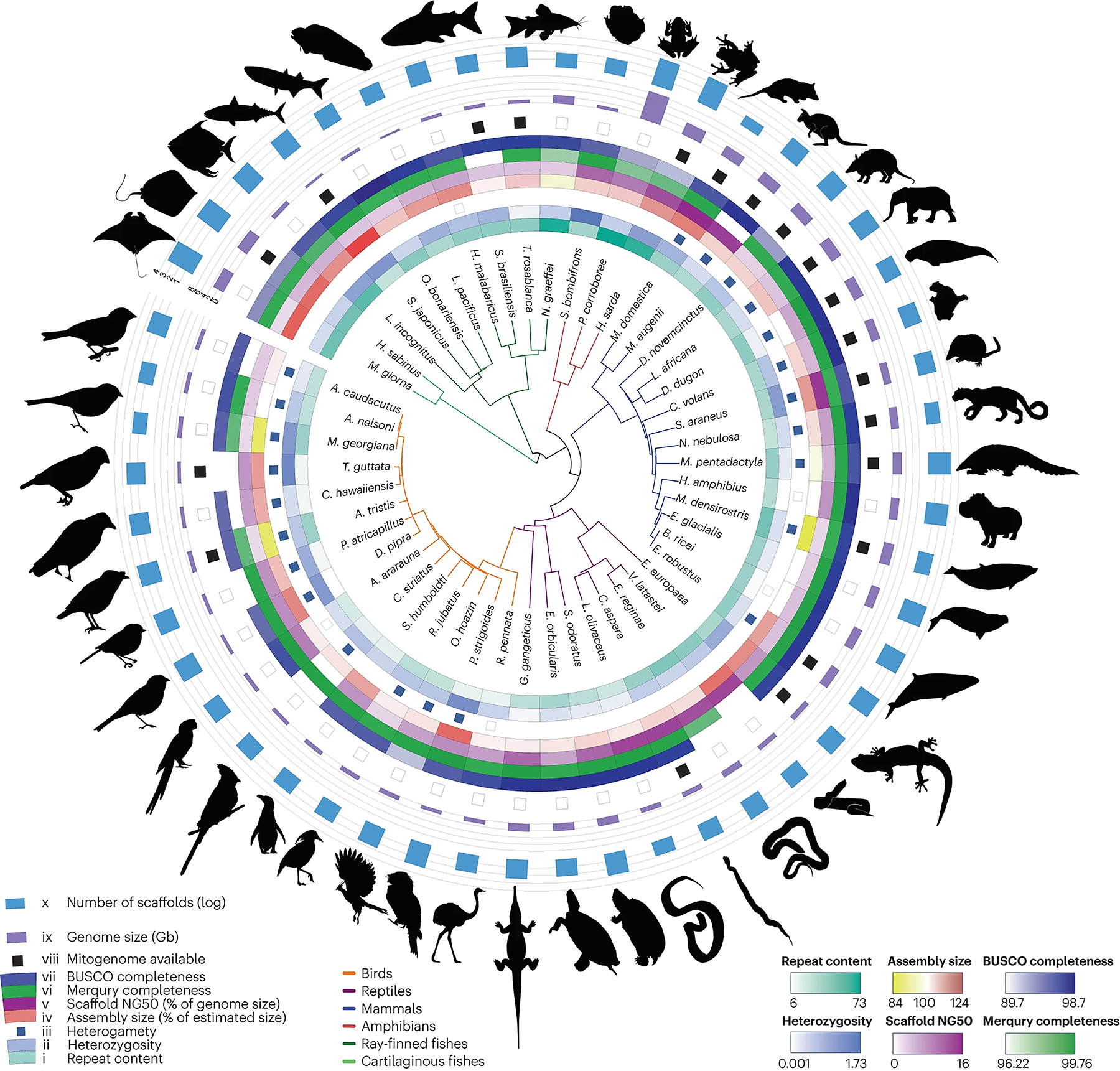
From the innermost circle to the outermost circle: (i) repeat content; (ii) heterozygosity; (iii) heterogamy: individuals with two identical sex chromosomes (white) or two different sex chromosomes (blue); (iv) assembly size in percentage of the genome size estimated by Genomescope; (v) scaffold NG50 in % of estimated genome size; (vi) Merqury completeness of both haplotypes; (vii) BUSCO completeness: presence of orthologous genes present and complete compared to the set expected in vertebrates; (viii) mitogenome assembled and available (black); (ix) genome size in gigabytes, with lines at 9, 2, 3, 4, 6 and 8 Gb; (x) number of scaffolds in log scale, with lines at 1 (10 scaffolds), 2 (100 scaffolds), 3 (1,000 scaffolds) and 4 (10,000 scaffolds).
