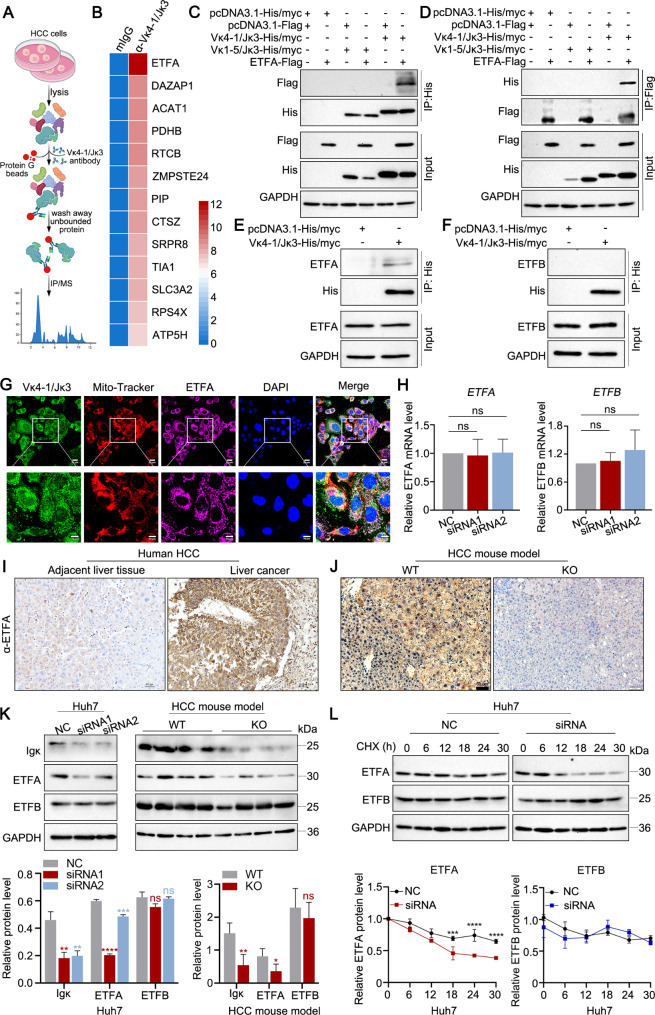
Fig. 5.
Hepatocyte-derived Igκ interacts with ETFA and maintains the stability of the ETFA protein. A Scheme displaying the procedure used for identifying the interacting protein of Igκ through Co-IP assay and mass spectrometry. B Heatmap showing the potential binding proteins of Vκ4-1/Jκ3-Igκ as determined by mass spectrometry. C - D The 293T cells were transfected with the indicated plasmids, and cell lysates were subjected to Co-IP using anti-His antibody magnetic beads (C) and anti-Flag antibody magnetic beads (D). E - F Total cell lysates from Huh7 cells overexpressing Vκ4-1/Jκ3-His/Myc were immunoprecipitated with anti-His antibody magnetic beads to detect the interaction between Vκ4-1/Jκ3-Igκ and endogenous ETFA (E) and ETFB (F). G The colocalization of Vκ4-1/Jκ3-Igκ (green) and ETFA (magenta) in mitochondria labelled with Mito-Tracker (red) in Huh7 cells was analysed by confocal immunofluorescence staining. Scale bars, 20 μm (upper) and 10 μm (lower). H The levels of ETFA and ETFB gene transcripts in Huh7 cells after Igκ knockdown were determined by qRT-PCR (n = 3). I Representative images of IHC for ETFA expression in liver cancer tissues and adjacent liver tissues (n = 9). Scale bars, 50 μm. J Representative IHC images of ETFA expression in liver tissues from WT and KO HCC mouse models (n = 5). Scale bars, 50 μm. K The protein levels of ETFA and ETFB in Huh7 cells with Igκ knockdown and liver tissues from WT and KO mice induced with DEN and CCL4 were detected by western blot analysis. Quantification of ETFA and ETFB protein levels relative to control GAPDH (Huh7: n = 3, HCC mouse model: n = 4). L Huh7 cells were transfected with Igκ siRNA or control siRNA and then treated with CHX (50 µg/mL) at the indicated time points. Western blot analysis was performed to evaluate ETFA and ETFB protein levels. The quantification of ETFA and ETFB expression levels is summarized (lower panel) (n = 3). The data are presented as the mean ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns, not significant