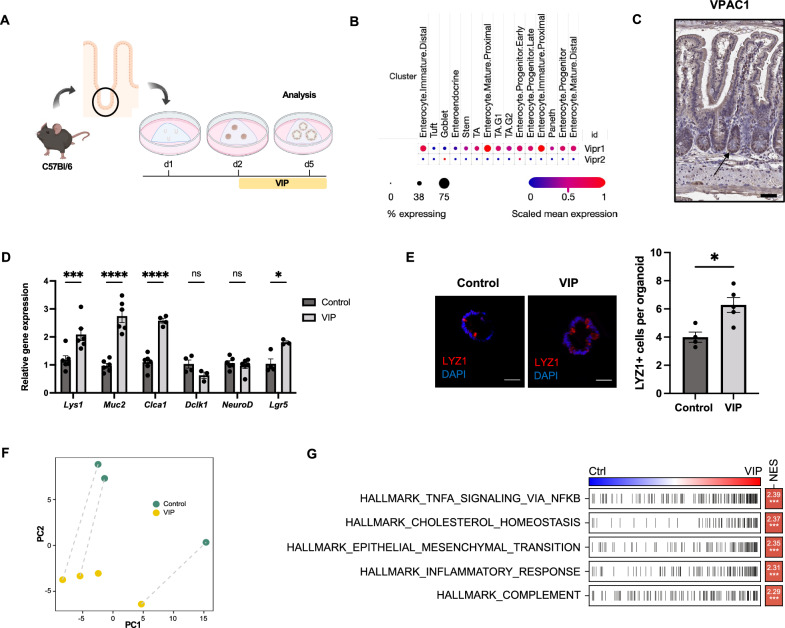
Fig. 1.
VIP modulates cellular differentiation towards a secretory phenotype. A Experimental setup and treatment scheme. B Expression of Vipr1 and Vipr2 genes in different epithelial compartments (based on single-cell RNA sequencing data set from [32]. C Immunohistochemical staining of VPAC1 in the murine small intestine (Scale bar = 100 µm). D qPCR analysis of differentiation markers of Paneth cells (Lyz1), goblet cells (Muc2, Clca1), tuft cells (Dclk1), enteroendocrine cells (NeuroD) and progenitor cells (Lgr5) upon VIP treatment. n = 3–6 per group. E Immunofluorescent analysis of LYZ1 staining in control and VIP-treated organoids (Scale bars = 50 µm). n = 4 (Control), n = 5 (VIP). F PCA of transformed count data from the bulk RNA sequencing of control and VIP-treated whole intestinal organoids. n = 3 (Control), n = 4 (VIP). G Hallmark gene-set enrichment analysis of differential gene expression in VIP-treated organoids; Ctrl = control, NES = normalized enrichment score. *p < 0.05, ***p < 0.001, ****p < 0.0001. Data are shown as means ± SEM, n = number of biological replicates. Statistical analysis was performed by two-way ANOVA with Šídák's multiple comparisons test (D) and paired Student’s t-test in (E)