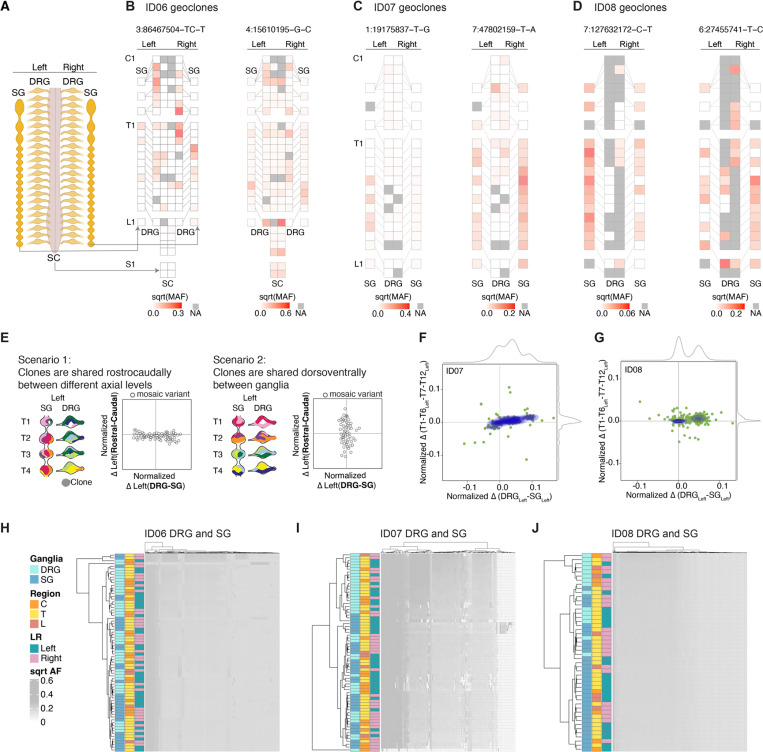
Figure 2.
DRGs or SGs share similar lineage relationships along the rostrocaudal axis but are clonally independent.
(A-D) Geographical maps of individual MV allelic fractions (AFs) (i.e. ‘geoclones’). Example geoclones in ID06 (B), ID07 (C), and ID08 (D) based upon hg38 reference genomic coordinate (at top). Colors: square root transformed mutant AFs (sqrt-MAF). NA: tissue not available.
(E) Models and contour plots of possible scenarios whereby MVs are shared between ganglia along the rostrocaudal axis but restricted dorsoventrally (scenario 1, left) or shared the dorsoventral axis while restricted rostrocaudally (scenario 2, right). Axes: normalized AF difference between rostral and caudal against that between DRG and SG. In scenario 1, most MVs deviate from the center along the x-axis but cluster at the center of the y-axis because of MVs are shared among rostral and caudal levels. In scenario 2, most MVs cluster around the center at the x-axis but not the y-axis since clones are more frequently shared dorsoventrally between DRG and SG, but not between different levels. Clones with genomic similarity are colored similarly.
(F-G) MV contour plots observed from ID07 (F) and ID08 (G) thoracic levels with the normalized difference in AFs between rostral (defined as T1-T6 levels) and caudal (defined as T7-T12 levels) (y-axis) plotted against the normalized difference between the DRG and SG (x-axis). Green dots: individual MVs. Blue contours: kernel density estimation of MV distributions. Grey curves: kernel density estimation along the respective axes. Only data for the left-sided ganglia are included in the plots.
(H-J) Hierarchical clustering heatmap with Manhattan distances using AFs of MVs from ID06 (H), ID07 (I), or ID08 (J). Note that DRGs and SGs predominantly clustered separately, whereas the left and right tend to intermix together, suggesting the lineage relationship between ganglia is not driven by their anatomical position but rather by their identity (DRG or SG). C: cervical, T: thoracic, L: lumbar.