Figure 3.
Concurrent single-nucleus MV genotyping with transcriptome analysis confirms the rostrocaudal clonal organization.
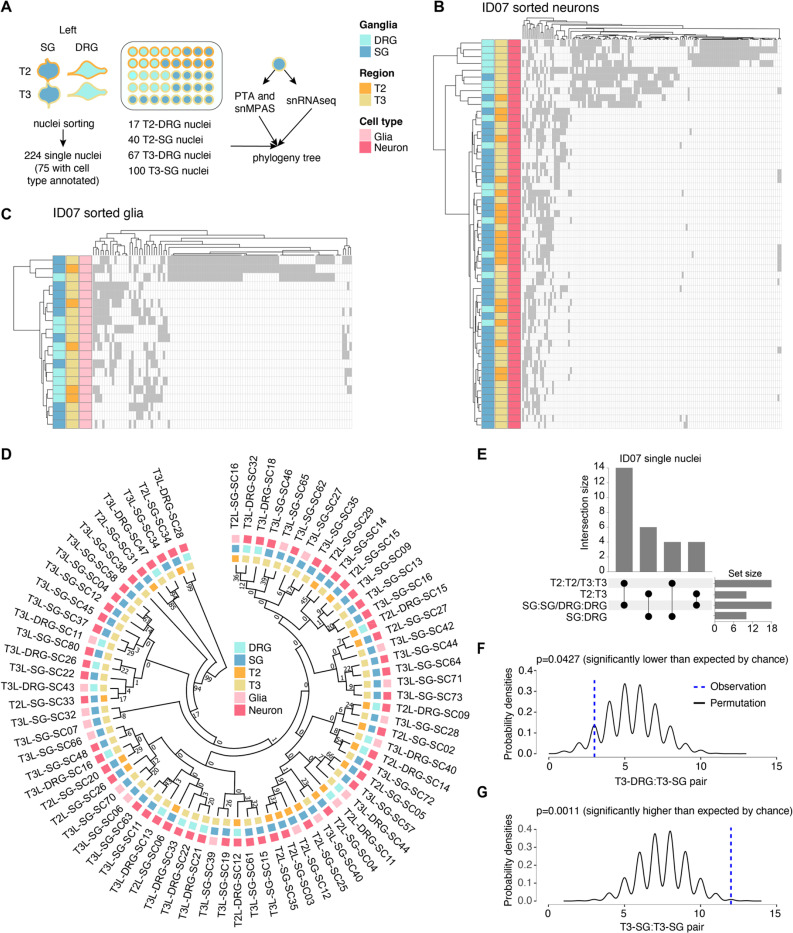
(A) Strategy for deconvolving the phylogenetic relationship of ganglia at single-cell resolution using ResolveOME (i.e. concurrent DNA amplification by primary template-directed amplification (PTA) and RNA profiling from the same nucleus). MVs genotyped by single-nucleus massive parallel amplicon sequencing (snMPAS) in 224 nuclei isolated from both the DRG and SG at T2 and T3 levels of donor ID07. Of these, cell type was inferred in 75 nuclei by snRNAseq.
(B-C) Hierarchical clustering with Manhattan distances from a total of 55 neurons (B) and 20 glia (C) from the DRG and SG at T2 and T3 levels of ID07. Note that DRGs and SGs predominantly clustered separately at both levels, suggesting the lineage relationship between ganglia is not driven by spinal level but rather by identity (DRG or SG).
(D) Phylogenic tree following 1,000 bootstrap replications based on the 184 MVs in 75 single nuclei with cell type information. Numbers at branches of the tree: bootstrap values supporting each edge.
(E) Upset plot showing number of terminal branches shared between type of ganglia (SG:SG, DRG:DRG or SG:DRG) and axial levels (T2:T2, T3:T3 or T2:T3).
(F-G) Number of terminal branches observed in the phylogeny tree in (D) (blue dashed line, observation) and distribution expected after 10,000 permutations (black line: permutation) for T3-DRG:T3-SG pair (p=0.0427, permutation test) (F) and T3-SG:T3-SG pair (p=0.0011, permutation test) (G).