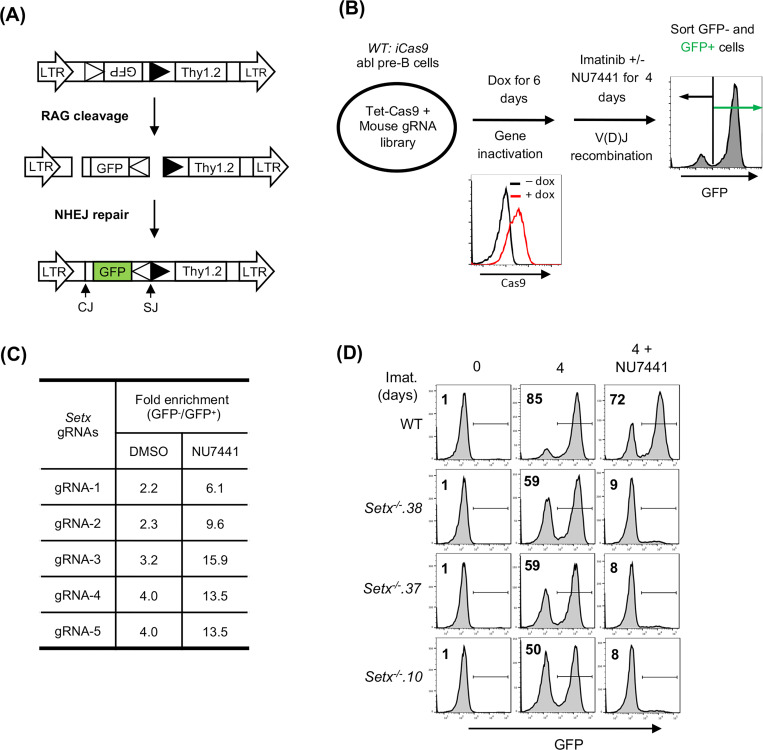
Figure 1: A Genome-wide CRISPR/Cas9 screen for identifying genes required for V(D)J recombination in DNA-PKcs inhibited abl pre-B cells.
(A) The schematic of the retroviral V(D)J recombination substrate pMG-INV. The open and filled triangles represent the RSSs. The retroviral LTRs, CEs and SEs upon RAG cleavage as well as CJs and SJs after NHEJ-mediated repair are indicated. The anti-sense GFP cDNA is inverted to the sense orientation upon completion of the recombination and its expression driven by the retroviral LTR (green). (B) The schematic diagram of a genome-wide CRISPR/Cas9 screen in WT abl pre-B cells for identifying genes important for V(D)J recombination upon DNA-PKcs inhibition with NU7441. (C) The fold enrichments of Setx gRNAs from screens conducted in DMSO and NU7441-treated WT abl pre-B cells (both also treated with imatinib). The fold enrichment is calculated as the ratio of the normalized count of a gRNA from GFP− cells over that from GFP+ cells. (D) Flow cytometric analysis for GFP expression in WT and three independently isolated Setx−/− abl pre-B cell lines with pMG-INV and treated with imatinib (imat.) in the presence or absence of the DNA-PKcs kinase inhibitor NU7441 for the indicated times. The percentages of GFP+ cells are indicated in the top left corners of the histograms.