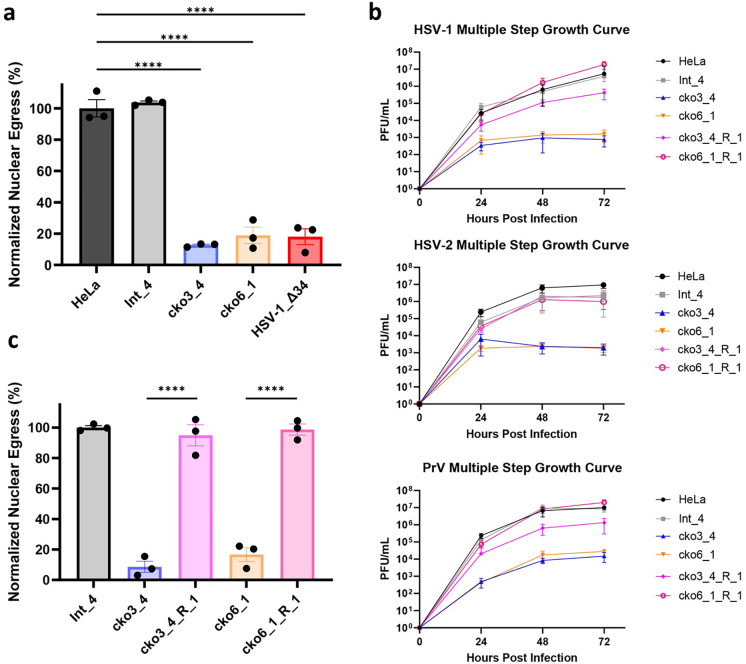
Figure 2. CLCC1 is essential for HSV-1 nuclear egress and HSV-1, HSV-2, and PrV replication.
a) Depletion of CLCC1 causes a defect in nuclear egress, measured by the flow cytometry nuclear egress assay. Single-clone CLCC1-KO (cko3_4 and cko6_1) or two control, HeLa and intergenic site targeting (Int_4) cell lines were infected with WT HSV-1 at an MOI of 5. As a positive control, HeLa cells were infected with HSV-1 ΔUL34 mutant virus, defective in nuclear egress, at an MOI of 10. Nuclear egress was measured at 24 hpi and normalized to HSV-1-infected HeLa cells. Each experiment had three biological replicates, each containing two technical replicates. Each data point represents a biological replicate. Bars represent mean values, and the error bars represent SEM. P < 0.0001 = ****. Significance was calculated using one-way ANOVA, with multiple comparisons. b) Multiple-step growth curves for HSV-1 on single-clone CLCC1-KO (cko3_4 and cko6_1), single-clone CLCC1-R (cko3_4_R_1 and cko6_1_R_1), or two control, HeLa and Int_4, cell lines. Cells were infected with HSV-1 at MOI of 0.1, and supernatants were titrated in Vero cells using plaque assay. The y-intercept is set to 10° at time 0 for visual clarity. Each symbol shows the mean of three biological replicates, each containing two technical replicates, and bars show SEM. c) Expression of CLCC1 in trans rescues the defect in nuclear egress, measured by the flow cytometry nuclear egress assay as in a). Single-cell CLCC1-KO (cko3_4 and cko6_1), single-cell CLCC1-R (cko3_4_R_1 and cko6_1_R_1), or control Int_4 cell lines were infected with HSV-1 at an MOI of 5. Nuclear egress was measured at 24 hpi and normalized to Int_4 signal. Each data point represents a biological replicate. Each experiment had three biological replicates, each containing two technical replicates. Each data point represents a biological replicate. Bars represent mean values, and the error bars represent SEM. P < 0.0001 = ****. Significance was calculated using one-way ANOVA, with multiple comparisons.