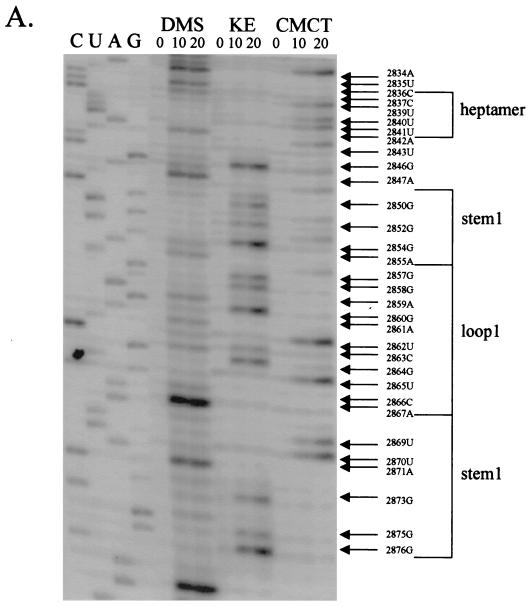
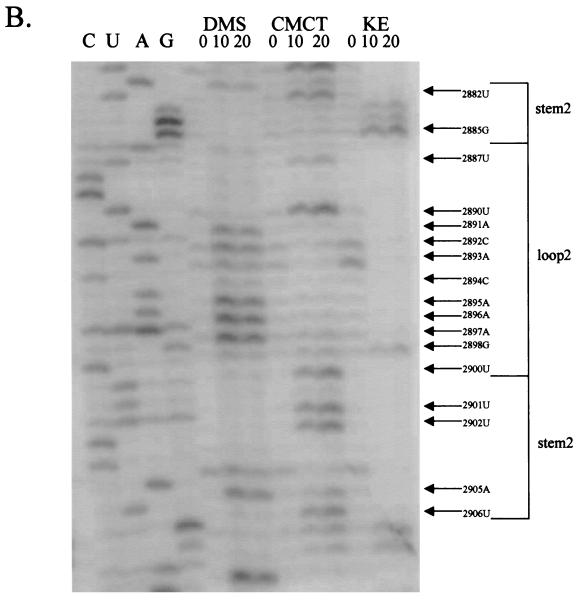
FIG. 5.
Chemical probings of the pC631(68)luc transcript and structural analysis of the 68-nt frameshift region by primer extension. Chemical modifications of A and C (by DMS), U (by CMCT), and G (by KE) were monitored by reverse transcription using end-labeled primers complementary to the nucleotides either 22 nt downstream of the last residue of stem-loop 1 (A) or 31 nt downstream of the last U residue of stem-loop 2 (B). The durations of incubation in each chemical reaction are indicated in minutes above each lane of gel electrophoresis. The left side of the gel labeled CUAG at the top represents the corresponding sequencing ladder of the cDNA. Arrows indicate transcriptional stops from primer extension representing the chemically modified bases in the treated RNA molecule, which migrate at a distance 1 nt short of that in the corresponding DNA ladder.