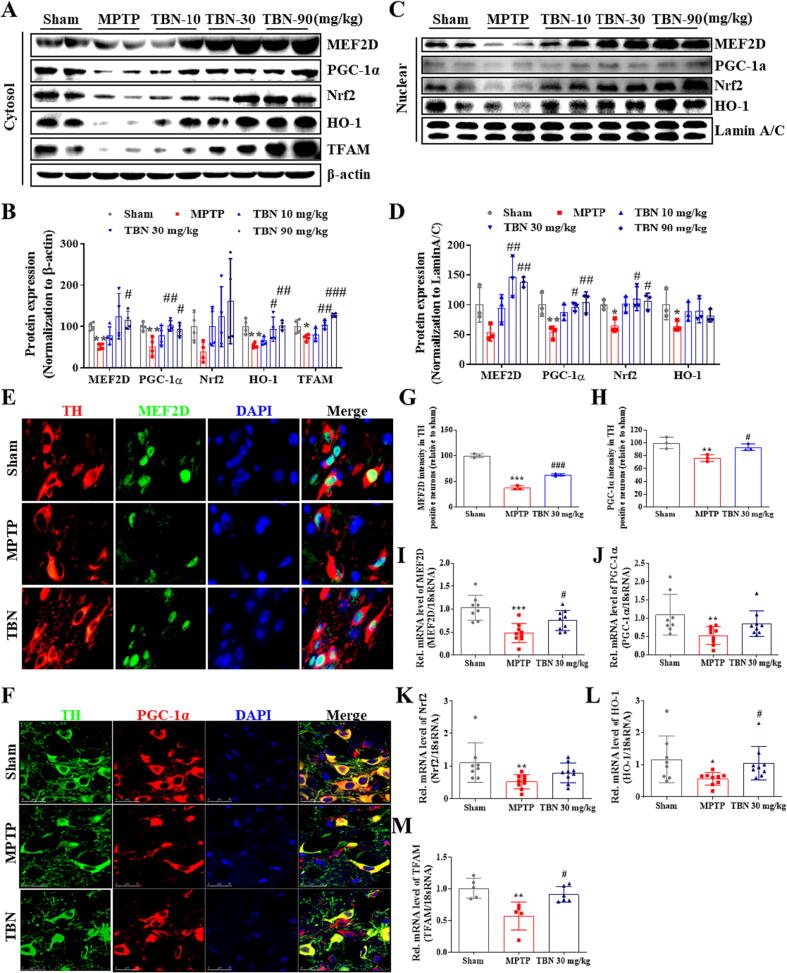
Fig. 6.
TBN activates PGC-1α/Nrf2 pathway in MPTP-induced PD mice. (A, B) Western blot for expression of MEF2D, PGC-1α, Nrf2, HO-1, and TFAM in the cytosol of SN tissue from MPTP mice. Data were mean ± SD of 4 mice per group. Data were analyzed using one-way Brown-Forsythe and Welch ANOVA test (MEF2D and Nrf2) or ANOVA followed by Dunnett’s multiple comparisons test (PGC-1α, HO-1, and TFAM). (C, D) Western blot for expression of MEF2D, PGC-1α, Nrf2, and HO-1 in the nuclei of SN tissue from MPTP mice. Data were mean ± SD of 3 mice per group. Data were analyzed using one-way ANOVA followed by Dunnett’s multiple comparisons test (MEF2D and PGC-1α) or Kruskal-Wallis test followed by Dunn’s multiple comparisons test (Nrf2 and HO-1). (E) Representative photomicrographs of co-immunofluorescence staining for TH (red), MEF2D (green), and DAPI (blue). Scale bars, 20 μm. (F) Representative photomicrographs of co-immunofluorescence staining for TH (green), PGC-1α (red), and DAPI (blue) in the SN of MPTP mice. Scale bars, 25 μm. (G, H) Fluorescence intensity of MEF2D and PGC-1α in the TH-positive neurons. Data were mean ± SD of 3 mice per group. Data were analyzed using one-way ANOVA followed by Dunnett’s multiple comparisons test. RNA from SN of PD animals was purified and converted into cDNA, and mRNA expression levels were determined using qRT-PCR. (I-M) Relative mRNA expression of (I) MEF2D, (J) PGC-1α, (K) Nrf2, (L) HO-1, and (M) TFAM in MPTP mice model. Data were mean ± SD of 5–9 mice per group. Data were analyzed using one-way ANOVA followed by Dunnett’s multiple comparisons test (I) or Kruskal-Wallis test followed by Dunn’s multiple comparisons test (J, K-M). *P < 0.05, **P < 0.01 and ***P < 0.001 versus sham group; #P < 0.05, ##P < 0.01 and ###P < 0.001 versus MPTP group. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)