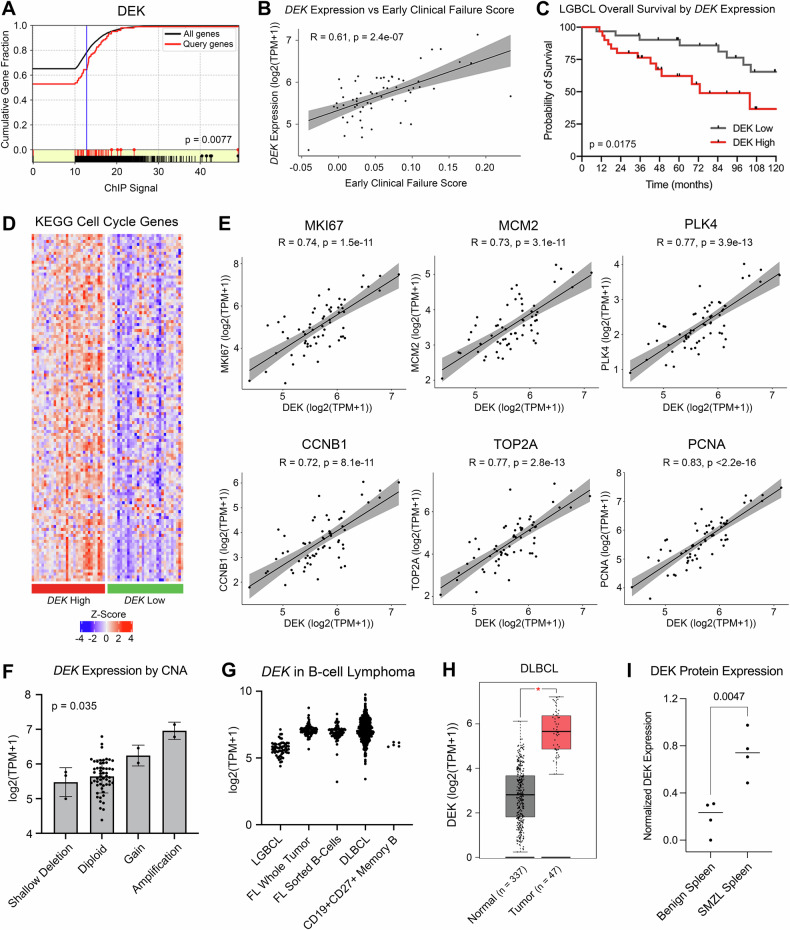
Fig. 1. DEK is associated with aggressive disease and is impacted by copy number alterations.
A ChIP signal of DEK protein as a regulator of early clinical failure signature genes identified by MAGIC TF analysis. B Correlation of DEK expression from LGBCL tumor RNA-seq data versus early clinical failure signature score. C Kaplan-Meier curves showing overall survival of patients expressing low vs. high levels of DEK. P-values were determined by log-rank test. D Gene expression of KEGG Cell Cycle genes from the Molecular Signatures Database. Heatmap shows z-scored log2(TPM + 1) values from LGBCL tumor RNA-sequencing data, with DEK high cases annotated in red and DEK low cases in green. E Correlation of DEK expression with expression of proliferation markers in RNA-seq data from LGBCL tumors. F DEK expression in LGBCL tumors by copy number status of DEK. G Log2(TPM + 1) expression of DEK across multiple B-cell lymphoma tumor types and healthy donor CD27 + CD19+ sorted memory B-cells. H DLBCL tumor versus normal DEK expression using the DLBC dataset from GEPIA. P-value was calculated in GEPIA using a one-way ANOVA with disease state as the variable for calculating differential expression. I Quantification of DEK expression from western blot analysis comparing benign spleens (n = 4) to SMZL spleens (n = 4). DEK expression levels were normalized to β-actin for each sample.