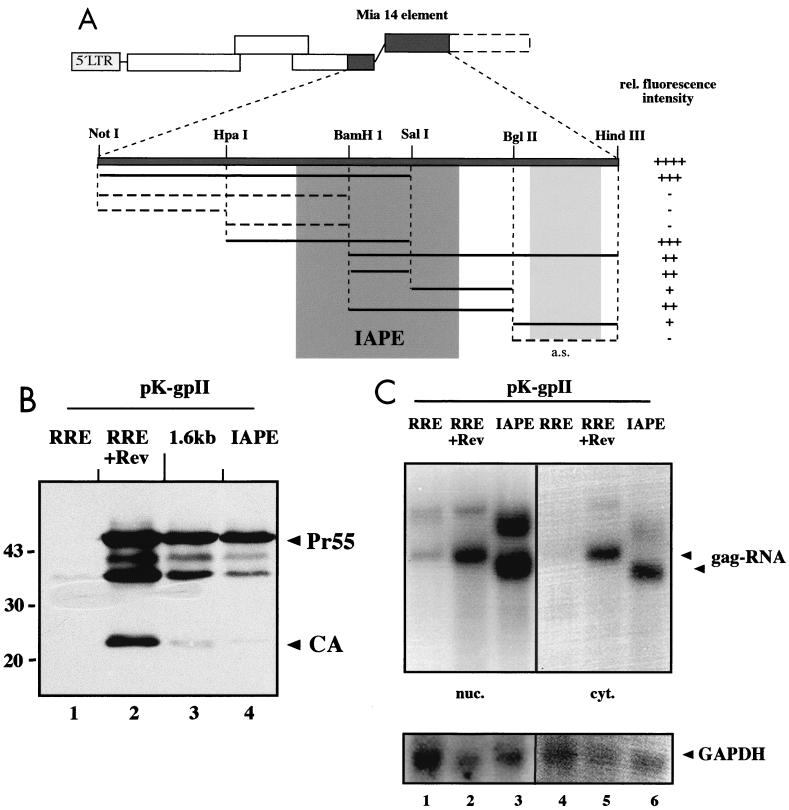
FIG. 3.
Mapping of the MIA14 expression element. (A) Schematic representation depicting the results of a functional analysis of various subfragments derived from the MIA14 element. The relevant restriction sites used for cloning of the subfragments are indicated. All subfragments were inserted into pK-gpII, and transiently transfected cells were analyzed for HIV-1 Gag expression. At least three independent experiments were performed for each construct. The relative (rel.) fluorescence intensity of each fragment is scored on the right. Subregions mediating Rev-independent Gag expression are shown as a solid line; those yielding negative results are depicted as a dashed line. Insertion of a subfragment in reverse orientation is termed antisense (a.s.). The central darkly shaded region corresponds to the region defined by structure prediction analysis (Fig. 4) and termed the IAPE. The lightly shaded region corresponds to a nonoverlapping sequence mediating weak Rev-independent Gag expression. (B) Western blot analysis of HeLa cells transfected with pK-R-gpII, with (+) or without Rev (lanes 1 and 2), pK-gpII-M (lane 3), or pK-gpII-IAPE (lane 4). HIV-1-specific products and marker protein were detected as described in the legend to Fig. 1. (C) Northern blot analysis of nuclear (nuc.; lanes 1 to 3) and cytoplasmic (cyt.; lanes 4 to 6) RNA preparations from HeLa cells transfected with plasmids pK-R-gpII, without and with (+) Rev (lanes 1 and 2 and 4 and 5), or pK-gpII-IAPE (lanes 3 and 6). The blot was hybridized with a radiolabeled probe directed against the gag-PR region present in all constructs. The arrowheads point to specific gag RNAs. The difference in migration results from size differences of the corresponding elements. The additional RNAs of lower mobility correspond to products derived from transcriptional readthrough into the hygromycin resistance transcription unit located 3′ of the polyadenylation signal. The same blot was subsequently stripped and reanalyzed with a probe specific for the cellular housekeeping GAPDH gene (bottom).