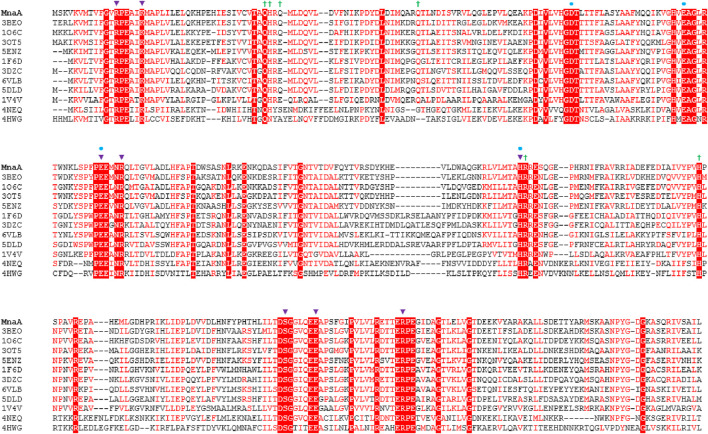
FIGURE 1.
Amino acid sequence alignment of non-hydrolyzing bacterial UDP-GlcNAc 2-epimerases currently published in the PDB (compare with Table 1), including PaMnaA (this study), showing conservation of catalytic and allosteric-site residues in PaMnaA. Red shading indicates residues that are 100% conserved across all epimerases while red font indicates highly conserved residues. The putative catalytic residues are marked with blue circles above the sequence, residues that bind the UDP-GlcNAc substrate are marked with purple triangles, and residues that bind the UDP-GlcNAc allosteric effector are marked with green crosses. 3BEO (Bacillus anthracis), 1O6C (Bacillus subtilis), 3OT5 (Listeria monocytogenes), 5ENZ (Staphylococcus aureus), 1F6D (E. coli), 3DZC (Vibrio cholerae), 6VLB (Neisseria meningitidis), 5DLD (Bacillus vietnamiensis), 1V4V (Thermus thermophilus), 4NEQ (Methanococcus jannaschii), 4HWG (Rickettsia bellii RML369-C) (Legg, 2022).