Abstract
Embryo implantation is crucial for ensuring a successful pregnancy outcome and subsequent child health. The intrauterine environment during the peri-implantation period shows drastic changes in gene expression and cellular metabolism in response to hormonal stimuli and reciprocal communication with embryos. Here, we performed spatial transcriptomic analysis to elucidate the mechanisms underlying embryo implantation. Transcriptome data revealed that lipid metabolism pathways, especially arachidonic acid–related (AA-related) ones, were enriched in the embryo-receptive luminal epithelia. Cyclooxygenases (COXs), rate-limiting enzymes involved in prostaglandin production by AA, were spatiotemporally regulated in the vicinity of embryos during implantation, but the role of each COX isozyme in the uterus for successful pregnancy was unclear. We established uterine-specific COX2-knockout (uKO) and COX1/uterine COX2-double-KO (COX1/COX2-DKO) mice. COX2 uKO caused deferred implantation with failed trophoblast invasion, resulting in subfertility with reduced pregnancy rates and litter sizes. COX1/COX2 DKO induced complete infertility, owing to abrogated embryo attachment. These results demonstrate that both isozymes have distinct roles during embryo implantation. Spatial transcriptome and lipidome analyses revealed unique profiles of prostaglandin synthesis by each COX isozyme and spatiotemporal expression patterns of downstream receptors throughout the endometrium. Our findings reveal previously unappreciated roles of COXs at the fetomaternal interface to establish early pregnancy.
Keywords: Endocrinology, Reproductive biology
Keywords: Eicosanoids, Reproductive biochemistry
Cox-1 and Cox-2 at feto-maternal interface differentially contribute to embryo spacing and invasion to establish early pregnancy by synthesizing unique PG species.
Introduction
Successful pregnancy requires appropriate conditions in the endometrium and healthy embryos (1, 2). Embryo implantation is a key process in which the embryos and uterine tissues dynamically interact (Figure 1A). In mice, the blastocyst arrives in the uterus in the early morning of day 4 of pregnancy (day 1 = plug-positive). Blastocysts hatch from their surrounding zona pellucida and have proper spacing between each embryo by the evening of day 4 (3). Embryos then interdigitate with the surface of the uterine luminal cells; this process is termed apposition. Once the blastocyst strongly attaches to the uterine epithelium on day 4 at midnight, the surrounding stromal cells initiate differentiation into decidual cells (decidualization). By the evening of day 5, trophoblast cells would have invaded the decidua by removing the surrounding luminal epithelial cell layer, eventually contributing to placenta formation. Any defects in the implantation and decidualization processes will have ripple effects on pregnancy outcomes; therefore, the interaction between the blastocyst and endometrium during early pregnancy must be tightly regulated (1). While accumulating studies using genetically engineered mice have revealed important genes that regulate embryo implantation (1, 2), it remains elusive how the expression of uterine genes changes spatiotemporally during the peri-implantation period.
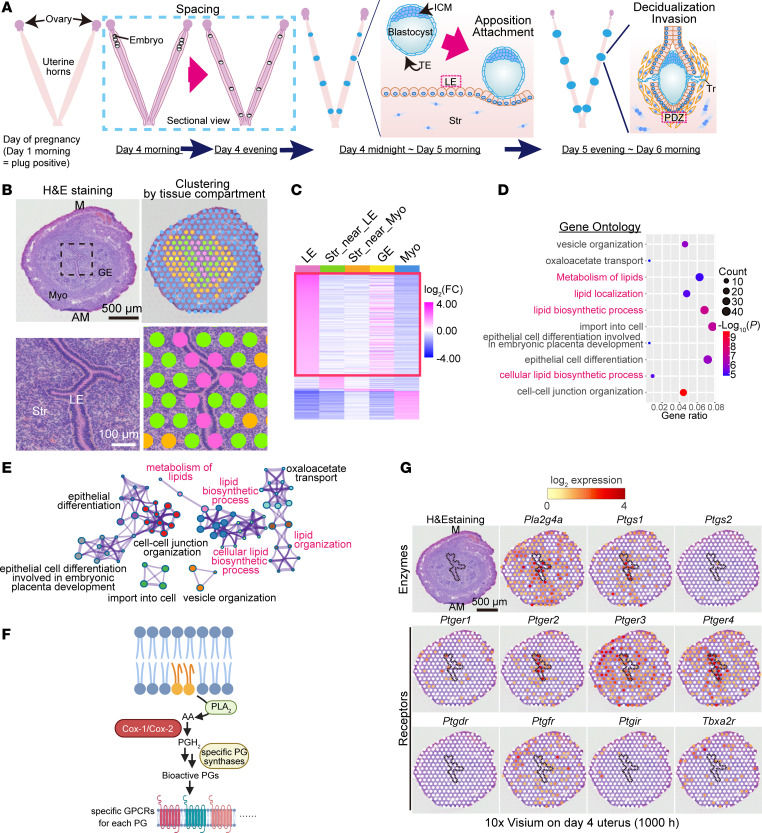
Figure 1. Spatial transcriptome revealing that lipid metabolism pathways are enriched in LE before embryo attachment.
(A) Schematic diagram of the process of embryo implantation in mice. ICM, inner cell mass; TE, trophectoderm; Tr, trophoblast; LE, luminal epithelia; Str, stroma; PDZ, primary decidual zone. (B and C) Spatial transcriptome using 10× Visium revealed LE-specific genes on day 4. In B, the area demarcated by a dashed line in the upper panels were magnified further in the lower panels. M, mesometrial pole; AM, anti-mesometrial pole; GE, glandular epithelia; Myo, myometria. Scale bars: 500 μm (upper in B) and 100 μm (lower in B). See also Supplemental Table 1. (D and E) Gene Ontology (GO) analyses using Metascape showed that LE-specific genes were enriched in lipid metabolism–related pathways. See also Supplemental Table 2. (F) Schematic diagram of PG synthesis from arachidonic acid (AA) and downstream GPCRs. (G) Spatial transcriptome of day 4 uteri showing unique expression patterns of PG synthesis–related enzymes (top) and receptors (middle and bottom). The area of LE is encircled by a dashed line. Scale bar: 500 μm. Day 4 (1000 hours) indicates 10 am on day 4 of pregnancy.
It is recognized that nonsteroidal antiinflammatory drugs (NSAIDs) impair early pregnancy events, including implantation and decidualization; hence, the intake of NSAIDs is prohibited in pregnant women (4). NSAIDs inhibit the enzymatic activities of cyclooxygenase 1 (COX1) and COX2, the rate-limiting enzymes in the synthesis of prostaglandins (PGs) from arachidonic acid (AA). PGs are bioactive lipids that exert their effects by activating specific cell membrane–spanning receptors (4–6). While COX1 and COX2 share structural features, with approximately 60% sequence homology, each enzyme shows a unique localization at both the tissue and subcellular levels (7). COX1 is considered the “housekeeping” COX, as it is expressed constitutively in most tissues, whereas COX2 expression is inducible by particular stimuli, such as inflammation and intracellular Ca2+ mobilization (5, 7). In cells, COX1 tends to localize to the endoplasmic reticulum, whereas COX2 preferably localizes to the perinuclear membrane. These differences in intracellular localization are considered to reflect the unique biological role of each isozyme. Indeed, in vitro studies have demonstrated that COX1 and COX2 have different substrate preferences, resulting in the synthesis of unique PG species (7). However, the functional differences between the 2 COXs in vivo have not been well clarified. In mouse uteri, cytosolic phospholipase A2α (cPLA2α) and COX1 are highly expressed in the luminal epithelium on day 4 in the morning. After embryo attachment, COX1 disappears, but COX2 colocalizes with cPLA2α at epithelial and stromal cells surrounding the embryos (8). These expression patterns suggest that COX1 and COX2 have differential roles in establishing early pregnancy. However, the differential functions of COXs in the uterus remain unclear (6) for the following reasons. First, in COX1-knockout (COX1-KO) uteri, no severe defects in early pregnancy were observed because COX2 compensated for the lost COX1 function (9, 10). Second, females lacking COX2 systemically are completely infertile due to defects in ovulation and fertilization (11); therefore, the uterine-specific roles of COX2 are obscure.
In this study, we conducted spatial transcriptomic analysis on mouse uteri and found that gene expression in lipid metabolism pathways, especially AA-related ones, are enriched at receptive luminal cells and primary decidual zones (PDZs), which directly contact to blastocysts. In addition, the spatiotemporal expression of COX1/COX2 and PG receptors was dramatically altered during the implantation processes. Lipidome analyses using liquid chromatography with tandem mass spectrometry (LC-MS/MS) and imaging MS also revealed that COX1 and COX2 uniquely play roles at different time points. To elucidate the differential roles of COX1 and COX2, we established uterine-specific COX2-KO (uKO) and COX1-KO/COX2-uKO double-KO (COX1/COX2-DKO) mice to investigate the functions of these 2 enzymes during pregnancy.
Results
Spatial transcriptome reveals AA/PG-related pathways in the fetomaternal interface during the embryo implantation period.
On the morning of day 4, blastocysts and endometria begin to physically communicate to establish embryo implantation (Figure 1A) (1). Luminal epithelia (LE) have direct contacts with embryos until they are eliminated after embryo attachment (Figure 1A) (12, 13), indicating critical roles for LE in embryo spacing and attachment.
To elucidate the landscape of signaling networks governing the functions of LE on day 4, we performed spatial transcriptome analysis using 10× Visium (Figure 1, B and C). Gene clustering according to the tissue compartment on the morning of day 4 revealed uniquely expressed genes in LE compared with that in other cell types (Figure 1C and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.181865DS1). We validated our 10× Visium data by immunostaining of cytokeratin 8 (CK-8; an epithelial marker), Ki67 (a proliferation marker; stromal cells should be stained on day 4; ref. 14), and Foxa2 (a glandular epithelium marker) (Supplemental Figure 1). Gene Ontology (GO) analyses showed that LE-unique genes were enriched in lipid metabolism–related pathways (Figure 1, D and E, and Supplemental Table 2). Notably, AA-related enzymes (e.g., Pla2g4a, Pla2g10, Lpcat3, and Acsl4) and receptors (Ptger2 and Ptger4) of PGs were detected (Supplemental Tables 1 and 2). Wang et al. (15) recently reported concentrated lipid metabolism in LE via single-cell RNA-sequencing (scRNA-seq) analyses. We reanalyzed their scRNA-seq data from mouse endometria on day 4 (3.5 day post coitus [dpc] in the original paper) and found that AA-related enzymes were highly expressed in LE (Supplemental Figure 2), supporting our 10× Visium data.
Transcriptome analyses enabled observation of the spatial expression of PG synthesis–related enzymes and downstream receptors on the morning of day 4 (Figure 1, F and G). In the mouse uterus, cPLA2α (encoded by Pla2g4a) is responsible for AA supply for PG synthesis (Figure 1F). Female mice lacking cPLA2α show deferred embryo implantation and crowded embryo spacing, resulting in shared placentae and reduced litter sizes (8). In agreement with this, our spatial transcriptome analysis showed prominent expression of Pla2g4a (cPLA2α) in LE with Ptgs1 (COX1), indicating crucial roles for cPLA2α and COX1 in these epithelia (Figure 1G). In contrast with COX1, the COX2 gene (Ptgs2) was poorly expressed in this milieu (Figure 1G). Each PG species produced by COXs triggers unique biological actions by binding to specific G protein–coupled receptors (GPCRs), although it remains unclear which PG receptor(s) is crucial for embryo implantation (4). The transcriptome data revealed that PGE2 receptors Ptger2, Ptger3, and Ptger4 were highly expressed on the morning of day 4, while Ptger1 showed a faint expression (Figure 1G). Notably, Ptger2 and Ptger4 were expressed on the luminal epithelium, where COX1 is also expressed (Figure 1G), suggesting possible roles of the COX1/PGE2 axis in implantation events in day 4 uteri. In contrast with the high expression of PGE2 receptors, other PG receptors, Ptgdr (a PGD2 receptor) and Ptgir (a PGI2 receptor), were faintly expressed (Figure 1G). PGD2 and PGI2 have another receptor, Ptgdr2 and Ppard, respectively (16, 17). Similar to Ptgdr, Ptgdr2 also showed poor expression in day 4 uteri (Supplemental Figure 3). Ppard encodes the nuclear receptor PPARδ (17). We found stromal expression of Ppard (Supplemental Figure 3). This observation agrees with that of a previous study demonstrating that PGI2 works in early pregnant uteri via PPARδ rather than GPCR (18); it is, however, unclear whether the PGI2/PPARδ axis works physiologically (19). The 2 remaining PG receptors, Ptgfr (a PGF2α receptor) and Tbxa2r (a thromboxane A2 [TxA2] receptor), were majorly found in the myometrial layer.
After embryo attachment, stromal cells undergo differentiation called decidualization (1) (Figure 1A). The decidua in the close vicinity of the attached blastocysts is the PDZ, which exhibits epithelial cell–like features with tight junctions (20, 21). The PDZ is considered important to protect embryos from maternal immune cells and pathogens instead of eliminated LE after embryo attachment (Figure 1A) (12, 13). In uteri with abnormal PDZ formation, embryo growth is compromised, with defective embryo invasion (21, 22), indicating the importance of the PDZ during the embryo invasion phase.
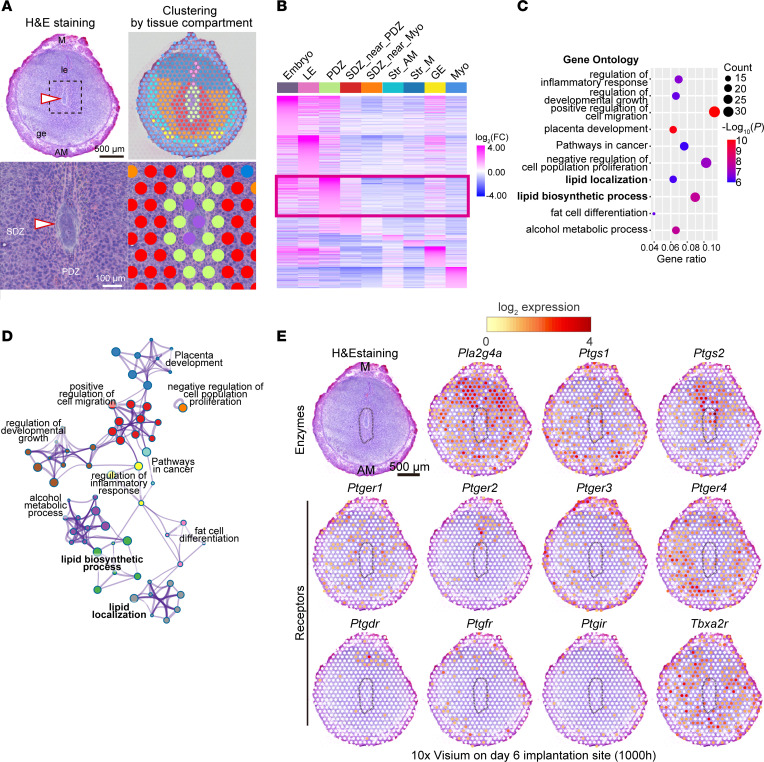
These results prompted us to investigate the gene network underlying PDZ functions in day 6 pregnant uteri (Figure 2). Our 10× Visium analyses revealed uniquely expressed genes in the PDZ (Figure 2, A and B, and Supplemental Table 3). The cell clustering was validated by immunostaining of Cdh1 (an epithelial marker) and Ptgs2 (a PDZ marker) as well as in situ hybridization of Bmp2 (a secondary decidual zone [SDZ] marker) (Supplemental Figure 4). Gene ontologies for these PDZ-specific genes showed high correlation with lipid metabolism–related pathways (Figure 2, C and D, and Supplemental Table 4). Notably, Ptgs2, which encodes COX2, was involved in most of the enriched GO pathways (Supplemental Table 4), indicating a critical role for COX2 in the PDZ. In agreement with our notion, Ptgs2 was highly accumulated in the endometrium in the vicinity of the attached embryo (Figure 2E), which was in contrast with the poor expression of this gene in day 4 uteri (Figure 1G). The possible contribution of COX2 was further assessed by spatial transcriptome analysis of each PG receptor (Figure 2E). Four PGE2 receptors as well as the PGD2 receptor were expressed in the vicinity of COX2 (Figure 2E). Notably, Ptger2 and Ptgdr were localized in the epithelium and stroma on the mesometrial side, whereas Ptger4 was evident in the SDZ and PDZ (Figure 2E). We detected faint expression of Ptgfr, Ptgir (Figure 2E), and Ptgdr2 (Supplemental Figure 5). In agreement with a previous report (18), Ppard was highly expressed throughout the decidua (Supplemental Figure 5). Tbxa2r showed spotty signals throughout the endometrium, but not in the PDZ (Figure 2E), indicating that it is expressed in endothelial cells, as the PDZ is avascular (21). Intriguingly, PGE2 receptors were widely distributed in uterine tissues, including the SDZ, whereas the PGD2 receptor was localized strictly near the embryo attachment site, where COX2 was also expressed (Figure 2E). This indicated that PGE2, but not PGD2, can be distributed throughout the endometrium.
Figure 2. Enriched lipid-related pathways in PDZ during the embryo invasion phase.
(A and B) Spatial transcriptome using 10× Visium revealed PDZ-specific genes on day 6. In A, the area demarcated by a dashed line in the upper panels was magnified further in the lower panels. M, mesometrial pole; AM, anti-mesometrial pole; LE, luminal epithelia; GE, glandular epithelia; Str, stroma; PDZ, primary decidual zone; SDZ, secondary decidual zone; Myo, myometria. Arrowheads indicate embryos. Scale bars: 500 μm (upper in A) and 100 μm (lower in A). See also Supplemental Table 3. (C and D) Gene Ontology (GO) analyses using Metascape identified PDZ-specific genes enriched in lipid metabolism–related pathways. See also Supplemental Table 4. (E) Spatial transcriptome of day 6 implantation sites, showing unique expression patterns of PG synthetic enzymes (top) and receptors (middle and bottom). PDZ is encircled by a dashed line. Scale bar: 500 μm. Day 6 (1000 hours) indicates 10 am on day 6 of pregnancy.
COX1 and COX2 differentially contribute to pregnancy outcomes.
To clarify the uterine roles of COX1 and COX2 during early pregnancy, we established COX2-uKO mice by crossing Ptgs2fl/fl mice with PgrCre/+ mice (Supplemental Figure 6), which do not exhibit ovarian defects, unlike systemic COX2-KO mice (11). We also established COX1/COX2-DKO mice by mating Ptgs1–/– mice with Ptgs2fl/fl PgrCre/+ ones to assess COX1 functions, while avoiding compensation by increased expression of COX2 in COX1 single KO (9).
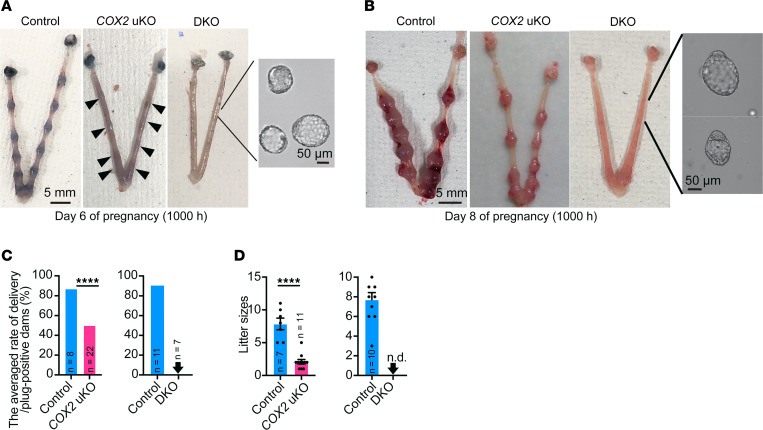
We first explored the physiological impact of COX1, which is highly expressed in luminal cells on the morning of day 4 (Figure 1G). Normally, after embryo attachment, embryo implantation sites can be visualized by an intravenous injection of blue dye due to increased vascular permeability in the endometrium in the vicinity of the attached embryos (23). However, COX1/COX2-DKO uteri never showed any implantation sites, with unattached blastocysts inside on day 6 of pregnancy (Figure 3A). This phenotype persisted even on day 8 of pregnancy (Figure 3B) when decidual cells reach terminal differentiation (24, 25). Eventually, the COX1/COX2-DKO mice were completely infertile, as they never delivered pups (Figure 3, C and D). This is in sharp contrast with the phenotype of COX2-uKO mice; the uteri from COX2-uKO mice had implantation sites both on days 6 and 8 of pregnancy, although blue dye reactions and sizes of each implantation site were smaller than those in the control (Figure 3, A and B). In addition, the average delivery rate in COX2-uKO females was 50%, with considerably reduced litter sizes, while it was 87.5% in the control (Figure 3, C and D). These results highlight the critical role of COX1 in the establishment of early pregnancy.
Figure 3. Defective pregnancy phenotypes in COX2-uKO and COX1/COX2-DKO mice.
(A and B) Representative photographs of uteri on day 6 (A) and day 8 (B). In COX1/COX2-DKO uteri, unattached blastocysts were flushed from the horns. Scale bars: 5 mm (uteri) and 50 μm (blastocysts). n = 3 per genotype. Day 6 (1000 hours) and day 8 (1000 hours) indicate 10 am on each day of pregnancy. (C and D) Pregnancy rates (C) and litter sizes (D) indicating subfertility and complete infertility in COX2-uKO and DKO mice, respectively. Arrowheads indicate zero value. n.d., not detected. Data are mean ± SEM. ****P < 0.0001 by Student’s t test. The number of samples is demonstrated on each bar.
We also observed that deletion of both COX1 and COX2 did not affect ovarian functions and uterine receptivity (Supplemental Figure 7). After ovulation and mating, high serum estrogen (E2) levels cause epithelial cell proliferation. By day 3, there is obvious stromal cell proliferation due to increased levels of progesterone (P4) secreted from the corpora lutea, while epithelial cell proliferation is terminated. This process, termed proliferation-differentiation switching (PDS), is critical for the establishment of an endometrium receptive for implantation-competent blastocysts (14). Abnormal PDS compromises embryo attachment and invasion (12, 26, 27); therefore, we suspected that PDS would be defective in COX1/COX2-DKO uteri. However, immunostaining for Ki67 revealed no significant defects in PDS in the mutant uteri on the morning of day 4 (Supplemental Figure 7A). In line with this finding, the expression levels of receptive marker genes in the endometria of COX2-uKO and COX1/COX2-DKO mice were comparable to those in control mice (Supplemental Figure 7B). Normal serum P4 levels further supported the notion that PDS occurred normally in both mutants (Supplemental Figure 7C). In summary, our results suggested a critical role of COX1 in uteri before embryo attachment, while both COX1 and COX2 are dispensable for the establishment of uterine receptivity.
Critical roles of COX1 in uterine PG synthesis and embryo spacing before embryo attachment.
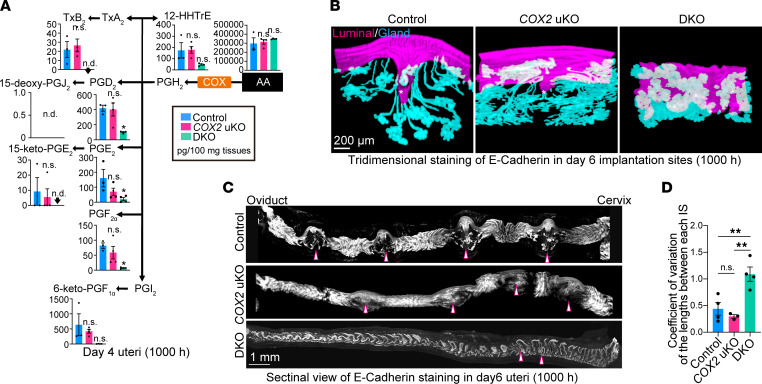
Next, we tested the enzymatic activities of each COX isozyme in day 4 uteri (Figure 4A). LC-MS/MS analysis found reduced levels of PGD2, PGE2, PGF2α, and TxB2 (a stable metabolite of TxA2) in COX1/COX2-DKO uteri (Figure 4A). In contrast, there were no obvious differences in the contents of any PGs between control and COX2-uKO mice (Figure 4A). These results were consistent with the fact that COX1, but not COX2, was highly expressed in the luminal epithelium on the morning of day 4 (Figure 1G) and failed embryo attachment was observed in COX1/COX2-DKO, but not COX2-uKO, uteri (Figure 3). According to the expression levels of each specific receptor (Figure 2E), PGE2, PGF2α, and TxA2, but not PGD2, might play roles in day 4 uteri.
Figure 4. COX1 is crucial for PG synthesis and embryo spacing before embryo attachment.
(A) Uterine PG concentrations on day 4, as analyzed by LC-MS/MS. n = 3 for control and n = 4 for COX2-uKO and COX1/COX2-DKO mice. n.d., not detected. Day 4 (1000 hours) indicates 10 am on day 4 of pregnancy. Data are presented as mean ± SEM. *P < 0.05 by 1-way ANOVA followed by Bonferroni’s post hoc test. n.d., not detected; n.s., not significant. (B and C) 3D images of luminal and glandular epithelia around embryo implantation sites (B) and whole uterine horns (C) on day 6 of pregnancy. Embryos are indicated by asterisks (B) or arrowheads (C). Scale bars: 200 μm (B) and 1 mm (C). At least 3 independent uteri were assessed for each genotype. Day 6 (1000 hours) indicates 10 am on day 6 of pregnancy. (D) The average coefficient of variation of the lengths between each implantation site (IS) was calculated from longitudinal scanning images shown in C. n = 3 for COX2-uKO and n = 4 for control and DKO. Data are presented as mean ± SEM. **P < 0.01 by 1-way ANOVA followed by Bonferroni’s post hoc test. n.s., not significant.
We then histologically evaluated pregnancy defects in each mutant line (Figure 4, B–D). A 3-dimensional (3D) view of the implantation site provides useful information for detecting anomalies in the implantation process and pregnancy maintenance (28, 29). After embryos attach to the luminal epithelium, the surrounding luminal layer forms crypts at the anti-mesometrial pole, accompanied by elongation of the glands. In uteri with failure of embryo attachment and decidualization, defects in this epithelial shaping have been found (29, 30). Previous studies have shown that embryo spacing occurs by the evening of day 4 (3, 31). Importantly, in Lpar3-KO uteri, which show crowded embryo implantation (32), abnormal embryo spacing is already observed on the evening of day 4 and sustained later (3, 31). As embryo implantation sites become evident on day 6 in healthy pregnancy (29), day 6 of pregnancy is the best time to determine whether embryo spacing, attachment, and decidualization are affected. We found poor crypt formation in COX2-deleted uteri, in contrast with the deeply evaginated luminal chamber and well-developed glands in control uteri (Figure 4B). COX1/COX2 DKO severely affected the epithelial structure surrounding blastocysts; DKO uteri showed neither crypt shaping nor gland elongation (Figure 4B) and were similar to the uteri before embryo attachment (28, 29). In addition to the epithelial layer abnormalities, blastocysts were located close to each other and to the cervical end in COX1/COX2-DKO uteri (Figure 4, B–D). Normally, blastocysts are evenly distributed in the uterine cavity during day 4 morning to afternoon (3, 31). Dysregulation of embryo spacing has also been observed in mice deficient in Lpar3 (encoding a GPCR for lysophosphatidic acid) or Pla2g4a (encoding cPLA2α), showing a shared placenta and embryo absorption (8, 32). Our observations suggested that COX1-derived PGs are essential for appropriate embryo spacing, while COX2 may play a role after embryo attachment.
COX2-derived PGD2 and PGE2 contribute to embryo chamber formation and embryo invasion.
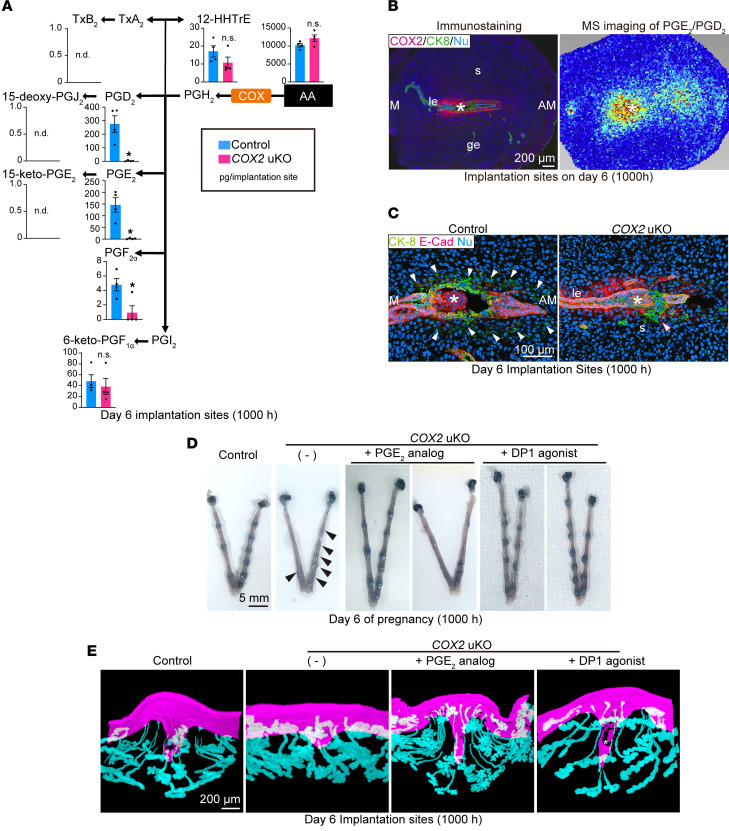
In contrast with the evident expression of COX1 before embryo attachment (Figure 1G), unique spatial expression of COX2 as well as PG receptors was found during the invasion phase (Figure 2E), leading us to examine the expression patterns of PGE2 and PGD2 synthetic enzymes in this milieu (Supplemental Figure 3). PGH2, synthesized from AA by COXs, is converted by specific PG synthases into various PG species that exert actions by binding to specific receptors (4, 7). In agreement with the widespread expression of PGE2 receptors in the endometrium (Figure 2E), PGE2 synthases (Ptges, Ptges2, and Ptges3) were widely detected in day 6 uteri (Supplemental Figure 8). In contrast, we observed local expression of hematopoietic PGD2 synthase (Hpgds) in the luminal epithelium (Supplemental Figure 8), which agreed well with the DP expression sites near the embryo attachment sites (Figure 2E). LC-MS/MS data also revealed PG expression in day 6 implantation sites, which were compared between control and COX2-uKO mice (Figure 5A). PGD2 and PGE2 levels were markedly reduced in COX2-uKO uteri. PGF2α levels were also decreased in COX2-uKO uteri, although they were substantially lower than those of PGD2 and PGE2. These results indicated that, on day 6, uterine PG synthesis depends entirely on COX2, in contrast with on day 4, where uterine PG synthesis largely depends on COX1 (Figure 4A). While we observed high expression levels of Txba2r (encoding a TxA2 receptor) in the SDZ (Figure 2E), the TxA2 metabolite was not detected (Figure 5A). It is possible that TxA2 in the circulating blood acts on the TxA2 receptor expressed in decidual blood vessels. These data suggested that COX2 is responsible for the massive production of PGE2 and PGD2 in day 6 uteri and contributes to the actions exerted by these PGs on day 6.
Figure 5. COX2-dependent PG production during the embryo invasion phase.
(A) PG concentrations in day 6 implantation sites, as analyzed by LC-MS/MS. n = 3 per genotype. Data are presented as mean ± SEM. *P < 0.05 by Student’s t test. n.d., not detected; n.s., not significant. Day 6 (1000 hours) indicates 10 am on day 6 of pregnancy. (B) Immunostaining of COX2 (left) and MS imaging of PGE2/PGD2 (right) on serial sections of day 6 implantation sites. M, mesometrial pole; AM, anti-mesometrial pole; LE, luminal epithelium; GE, glandular epithelium; S, stroma. Asterisks indicate embryos. Scale bar: 200 μm. Day 6 (1000 hours) indicates 10 am on day 6 of pregnancy. Three independent sections were assessed for immunostaining and MS imaging. (C) Coimmunostaining of CK-8 (a marker of trophoblasts and epithelia) and E-cadherin (a marker of epithelia) showing defective embryo invasion in COX2-uKO mice on day 6. Asterisks indicate embryos; arrowheads indicate invading trophoblasts. Scale bar: 100 μm. Three independent sections were assessed for each genotype. Day 6 (1000 hours) indicates 10 am on day 6 of pregnancy. (D) Representative images of day 6 uteri from control and COX2-uKO mice with or without PGE2 analog/DP1 agonist administration. Arrowheads indicate faint implantation sites. Scale bar: 5 mm. n = 3 per group. Day 6 (1000 hours) indicates 10 am on day 6 of pregnancy. (E) 3D images of luminal and glandular epithelia around embryo implantation sites from control and COX2-uKO mice with or without PGE2 analog/DP1 agonist administration on day 6 of pregnancy. Asterisks indicate embryos. Scale bar: 200 μm. Three independent uteri were assessed in each group. Day 6 (1000 hours) indicates 10 am on day 6 of pregnancy.
Imaging MS provided profound insights into the localization of PGE2 and PGD2 in the decidua (Figure 5B). Evident signals of PGE2 and PGD2 aligned along the luminal layers around the attached embryo, which was consistent with COX2 expression, as indicated by immunostaining (Figure 5B) and spatial transcriptome analysis (Figure 2E). Based on the spatial expression profiles of the specific synthases and receptors for PGE2 and PGD2 (Figure 2E and Supplemental Figure 8), PGD2 appears to function locally, within the vicinity of the embryos, whereas PGE2 likely acts after spreading out toward the anti-mesometrial pole of the decidua (Figure 5B). Taken together, these results demonstrated that COX1 and COX2 differentially contribute to PG synthesis in each phase of pregnancy, and specific receptors with unique localizations exist for each PG species.
The findings of the spatial transcriptome and lipidome analyses raised the question of how uterine COX2 influences implantation processes, especially on day 6 of pregnancy (Figure 5C). After embryo attachment, cell-cell adhesion between luminal epithelial cells in the vicinity of the embryo becomes weaker, contributing to subsequent trophoblast invasion into the decidua (13). Embryo invasion was evident on the morning of day 6, when CK-8–positive trophoblasts laterally infiltrated the deciduae (Figure 3C) (13, 26). However, in contrast with control uteri, COX2-uKO uteri showed defective epithelial cell removal, accompanied by anomalous trophoblast invasion (Figure 5C), indicating the importance of COX2 in this process. The spatial gene expression analyses and PG synthesis–related gene expression profiling allowed us to further investigate the individual roles of PGE2 and PGD2 in the uterus during the embryo invasion phase (Figure 5, D and E). To examine the effects of each PG species in COX2-dependent decidual functions, we subcutaneously injected COX2-uKO females with a PGE2 analog or a DP1 (PGD2 receptor) agonist on the morning of day 5. Compared with the control (no injection), both the PGE2 analog and the DP1 agonist enhanced blue reactions in the embryo implantation sites (Figure 5D). We also observed 3D structures of the implantation sites in each group, as a flawed formation of implantation chamber is related to failed embryo invasion, owing to defects of morphological changes in uterine epithelia (21, 33). As shown above (Figure 3A), shallow implantation chambers were found in the COX2-deleted milieu (Figure 5E). However, this abnormality was recovered upon administration of the PGE2 analog or DP1 agonist, resulting in deeply evaginated LE with elongated glands (Figure 5E). These results indicated the critical roles of PGE2 and DP1 signaling in the formation of implantation chambers. These results demonstrated the role of the COX2/PGE2/PGD2 axis in the establishment of appropriate implantation chambers, facilitating embryo invasion of the decidua.
Discussion
Embryo implantation, which involves intimate reciprocal communication between the endometrium and embryo, is key for pregnancy maintenance and healthy birth (1). Recently, spatial transcriptome analyses of fetomaternal interfaces have been conducted in humans and mice (34–36), providing previously unknown insights into how maternal tissues communicate with embryos in early pregnancy. However, these previous studies utilized endometria with fully invaded trophoblasts; therefore, the spatiotemporal gene landscapes working during the peri-implantation phase remain unclear. In this study, we analyzed mouse uterine tissues immediately before and after embryo attachment, showing that lipid metabolism–related pathways were enriched in LEs and PDZs directly in contact with embryos upon embryo spacing, attachment, and invasion.
It is well recognized that the intake of NSAIDs during early pregnancy compromises embryo implantation, resulting in embryo absorption (4). COX1 and COX2, the targets of NSAIDs, are the rate-limiting enzymes of PG synthesis. COXs utilize AA provided by PLA2s to produce PGG2, which is immediately converted to PGH2 in cells (5–7). PG synthases specific for each PG are responsible for the generation of bioactive PGs from PGH2, triggering GPCR signaling in an autocrine or paracrine manner. In vitro studies have demonstrated that PLA2s, COXs, and specific PG synthases show preferential coupling (7). Uteri of pregnant animals are remarkable in vivo models to investigate the differential roles of COX1 and COX2. Both enzymes are spatiotemporally expressed in the uterus during the peri-implantation period, with mutually exclusive expression patterns (8, 37). A previous study using reciprocal COX1- and COX2-knockin mice revealed reduced fertility, with defective embryo implantation (38); the authors prepared mice with knockin alleles of COX1 at the COX2 loci (COX1 > COX2) as well as reciprocally knocking in alleles of COX1 and COX2 (Reversa). Intriguingly, Reversa females exhibited a more severe phenotype during embryo implantation than COX1 > COX2 females, indicating that both COX1 and COX2 contribute to early pregnancy outcomes. These results suggested that each COX enzyme has a unique and specific role in the pregnant uterus.
This study revealed that COX1 is responsible for the synthesis of PGE2, PGD2, PGI2, and TxA2 in the receptive endometrium, while COX2 mainly produces PGE2 and PGD2 during embryo invasion. It remains unclear exactly how each enzyme is related to the synthesis of unique PGs in vivo, but our observations in day 6 uteri indicated that COX2 may be related to hematopoietic PGD2 synthase (Hpgds). Intriguingly, PGE2-producing enzymes did not colocalize with COX2 during the invasion phase. As COX2-produced PGH2 is unstable, it has to be immediately converted into each PG by specific synthases. Therefore, the question arises as to how COX2 couples with PGE2 synthases in early pregnant uteri. As shown in a recent study (39), it is possible that PGs, including PGH2, are transferred to cells expressing specific PG synthases and receptors via extracellular vesicles.
Our current study also demonstrates previously unappreciated roles of COX1 in early pregnancy. Notably, abnormal embryo spacing was found in COX1/COX2-DKO uteri, but not in COX2-uKO uteri, indicating the involvement of COX1-producing PGs in this process. However, which PG species play a major role remains elusive. There are reports showing that muscle contractions contribute to embryo spacing (40, 41), indicating that PGs might induce muscle contraction by being distributed throughout the endometria, while COX1 is expressed in LE. Indeed, we observed reduced production of 6-keto-PGF1α (a metabolite of PGI2) and the absence of TxB2 (a metabolite of TxA2) in COX1/COX2-DKO uteri on day 4, in addition to myometrial expression of their receptors. In contrast, accumulating studies using 3D imaging have shown that planar polarity in epithelia is another important factor regulating embryo spacing (28, 29). Therefore, we cannot ignore the possibility that PGs synthesized by COX1 work on LE. As single deletion of each PG receptor does not influence uterine functions (4), multiple PG receptors might cooperate to regulate embryo spacing. Another evident phenotype in COX1/COX2-DKO is flawed embryo attachment accompanied with abolished blue dye reactions. As embryo attachment induces increased vascular permeability in the endometria, embryo-attached sites can be visualized by the injection of blue dye (23). Matsumoto et al. reported that systemic deletion of COX2 causes reduced angiogenesis in the endometria (42), indicating possible roles of PGs in endothelia or immune cells. As previously reported (43), endometria and endothelia can have opposite effects against pregnancy outcomes, and thus it would be intriguing to see whether endothelial cell–specific deletion of COX1 or COX2 has any roles in early pregnancy.
Interestingly, defective implantation in COX2-uKO mice was rescued by a single administration of a DP1 agonist. While PGE2 can trigger various G protein signaling pathways, including Gs activation, via 4 PGE receptor subtypes, the DP1 agonist only triggers Gs signaling via the DP1 receptor; thus, PGE2 and the DP1 agonist may share Gs signaling activation (4). Our result suggests a critical role for the increased cAMP in decidualization; indeed, both Ptger2 and Ptgdr were expressed in luminal epithelial cells in day 6 uteri (Figure 5B). A previous study reported that the the COX2/PGE2 axis contributes to decidual reactions by upregulating stromal cAMP levels downstream of EP2 and EP4 (44). In addition, cAMP is required in primary culture of human uterine stroma to support decidualization in vitro (45). These findings suggest that PGE2 and PGD2 may exert their functions mainly via Gs-coupled receptors. This would also explain how COX2/PG signaling elicited a transformation in epithelial morphology (Figure 5, D and E). Recently, Ishihara et al. reported that PGE2/EP2/cAMP signaling is involved in YAP-induced cell competition in epithelial Madin-Darby canine kidney cells. In epithelial cells in which YAP was activated by physical stimuli, COX2 was induced to produce PGE2, which induced E-cadherin internalization only in PG-producing cells and adjacent cells in an EP2/cAMP-dependent manner, thereby eliminating YAP-activated cells (46). As EP2 and DP1 receptors share Gs activation, it is possible that uterine epithelia transform their shapes via the PGE2/PGD2/EP2/EP4/DP1/cAMP axis. As a previous study showed prominent localization of YAP in LE as well as PDZ cells, which exhibit epithelial cell–like features (21), such a system may also work in the endometrium during early pregnancy. Exogenous administration of PGE2 or PGD2 helped recover decidual reactions, but failed to rescue embryo growth in COX2-uKO mice. If a cell-competitive mechanism occurs sequentially and away from the embryo attachment site during epithelial breakdown, it requires the formation of a local PG concentration gradient, which may be difficult to achieve with exogenous PG administration. Mouse models with spatiotemporal knockout or induction of PG-related enzymes would be useful to further investigate this in the future.
In conclusion, the current study showed that lipid metabolism pathways, especially 2 COX isoforms (COX1 and COX2), differentially contribute to peri-implantation events in the mouse uterus in a spatiotemporal manner. This study revealed that PG species perform unique functions downstream of COX1 and COX2 to facilitate embryo spacing, attachment, and invasion. As COX1 and COX2 are also found in the human endometrium during implantation (47), the mechanism found in mice may be conserved among species, including humans.
Methods
Sex as a biological variable.
We utilized female mice for our analyses, as pregnancy occurs only in females.
Mice.
Ptgs2 (COX2)-loxP/loxP, Pgr-Cre, and Ptgs1 (COX1)-KO mice were used in this study. Ptgs2fl/fl mice (48) were provided by Harvey R. Herschman (UCLA, Los Angeles, California, USA). Ptgs1-KO mice (49) were supplied by Taconic Biosciences. Pgr-Cre mice (50) were provided by Francesco J. DeMayo (National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina, USA) and John P. Lydon (Baylor College of Medicine, Houston, Texas, USA). Ptgs2-loxP/loxP females were crossed with Pgr-Cre males to generate COX2-uKO mice. COX2-uKO males were further mated with COX1-KO females to establish COX1/COX2-DKO mice. All mice were housed under a 12-hour light/12-hour dark cycle with lights on from 08:00 to 20:00 hours in humidity-controlled rooms at 22°C at the University of Tokyo Animal Care Facility according to the institutional guidelines for the use of laboratory animals.
Evaluation of pregnancy outcomes.
To examine pregnancy outcomes, COX2-uKO, COX1/COX2-DKO, or COX2fl/fl (control) female mice were mated with C57BL/6N fertile male mice, as reported in a previous study (12, 30, 51). The day when the vaginal plug was detected was considered as day 1 of pregnancy. Pregnant mice were euthanized by cervical dislocation on designated days of pregnancy for the evaluation of pregnancy phenotypes and sample collection. On day 4, one uterine horn was flushed with saline to confirm the presence of blastocysts. Embryo attachment sites were observed as blue bands soon after intravenous injection of 1% solution of Chicago blue dye (Sigma-Aldrich) in saline on days 5 and 6. When no embryo attachment sites were observed as of day 5, both uterine horns were cut and flushed with saline to collect embryos.
DNA/RNA extraction.
DNA and RNA were extracted from homogenized tissues using TRI Reagent (Molecular Research Center Inc.) according to the manufacturer’s protocol. The concentrations of each sample were tested using a NanoDrop spectrophotometer (Thermo Fisher Scientific). The complementary DNA was synthesized from the extracted RNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO).
PCR to check genomic deletion of COX2.
DNA collected from uterine tissues was used as a template. One hundred nanograms of DNA was amplified using the primers 5′-AATTACTGCTGAAGCCCACC-3′ and 5′-GAATCTCCTAGAACTGACTGG-3′ and rTaq (Takara), with 35 cycles of 98°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. The PCR products were electrophoresed in a 2% agarose gel to detect a band shift.
Quantitative reverse transcription PCR.
qPCRs were run using THUNDERBIRD SYBR qPCR Mix (TOYOBO). The housekeeping gene Actb was used for internal standardization of mRNA expression. Relative expression levels were determined by the ΔΔCt method (52). The following primers were used: Ptgs1 5′-TCCAGGAGCTCACAGGAGA-3′ and 5′-GTCACCACCGAACGTGCT-3′; Hdc 5′-GCCCATCTGTGCCAGTGAGGGA-3′ and 5′-GAAAGCGCCGGCTCAAGGGG-3′; Ihh 5′-GAGAACACGGGTGCCGACCG-3′ and 5′-CAGCGGCCGAATGCTCAGACT-3′; Muc1 5′-GTGCCAGTGCCGCCGAAAGA-3′ and 5′-CCGCCAAAGCTGCCCCAAGT-3′; Actb 5′-TGTTACCAACTGGGACGACA-3′ and 5′-GGGGTGTTGAAGGTCTCAAA-3′.
Immunofluorescence.
Frozen sections (thickness, 12 μm) were prepared by CM1950 cryostat (Leica) and used for immunofluorescence. After fixation in 4% paraformaldehyde in PBS, the sections were incubated with primary antibodies against Ki67 (SP6, Thermo Fisher Scientific; 1:300), COX2 (160106, Cayman; 1:300), E-cadherin (24E10, Cell Signaling Technology; 1:300), and CK-8 (TROMA-I, DSHB; 1:300). Signals were detected using Alexa Fluor 555–conjugated anti–rabbit IgG (A21428, Thermo Fisher Scientific; 1:500), Alexa Fluor 488–conjugated anti–rat IgG (A11006, Thermo Fisher Scientific; 1:500), and 4′,6-diamidino-2-phenylindole (DAPI) (Dojindo, 1:500). Images were acquired using an AXR confocal microscopy system (Nikon).
FISH.
We performed FISH as previously reported (21, 53), with some modifications. Briefly, frozen sections with 12 μm thickness were attached on Superfrost Plus Microscope Slides (Thermo Fisher Scientific). After fixation and acetylation, the sections were hybridized overnight at 55°C with a DIG-labeled Bmp2 antisense probe. Following probe washing, the sections were treated with an anti-DIG antibody-POD (11207733910, Roche; 1:200), followed by Alexa Fluor 488–conjugated tyramide (B40953, Invitrogen). Nuclei were counterstained with DAPI. Images were acquired using an AXR confocal microscopy system.
LC-MS/MS.
Pregnant uteri were collected in the morning of days 4 and 6 and processed for LC-MS/MS–based lipidome analyses, as previously described (54). Briefly, collected tissues were snap-frozen and homogenized in methanol, and lipids were extracted by single-phase extraction. Lipidome analysis was performed using the ACQUITY UPLC system (Waters) coupled with quadruple time-of-flight/MS (TripleTOF 5600 or 6600, SCIEX).
Imaging MS analysis.
Fresh-frozen sections (12 μm thick) of day 6 implantation sites were attached to a conductive indium tin oxide–coated glass slide (Matsunami Glass Industries). For on-tissue derivatization prior to matrix coating, Girard T reagent (G900-100G, Merck), which converts the ketone groups of PGE2 and PGD2 into the positively charged hydrazone, was prepared at 10 mg/mL in 20% acetic acid (Fujifilm Wako Pure Chemical Corporation). One milliliter of Girard T reagent was sprayed onto each glass slide using an artist’s airbrush (PS-270, GSI Creos). The sections were then airbrushed with DHB matrix coating (490-79-9, Bruker, 50 mg/mL in 80% ethanol). Imaging MS with ion mobility separation was performed using a matrix-assisted laser desorption/ionization-time-of-flight instrument (timsTOF flex, Bruker Daltonics). The tissue surface was irradiated with a YAG laser (500 laser shots/pixel) in the positive ion detection mode and the spot-to-spot center distance was set to 35 μm. The ion mobility conditions were optimized to separate the ions of Girard T–derivatized PGE2 and PGD2 by measuring reference standards. The obtained MS spectra were reconstructed into MS images using the Scils Lab software (Bruker Daltonics).
Spatial transcriptome analysis.
Spatial transcriptome analysis was performed using 10× Visium (10× Genomics) according to the manufacturer’s protocol. Briefly, frozen sections (10 μm thick) were mounted on gene expression slides and sent to KOTAI Biotechnologies for subsequent procedures. After reaction with Proteinase K for 40 minutes, the sections were hybridized with spatial tags on the slides and reverse transcribed in situ. The synthesized cDNAs were subjected to RNA-seq using DNBseq (MGI), producing 300 million reads/sample.
Raw FASTQ files and microscope slide images for each sample were processed using the Space Ranger software (version 1.1, 10× Genomics) with the spaceranger count pipeline, using STAR with default parameters for aligning reads against the mouse reference genome mm10 refdata-gex-mm10-2020-A. This pipeline uses the Visium spatial barcodes to generate a feature-spot matrix of unique molecular identifier counts. To visualize genes of interest, we used Loupe Browser (10× Genomics) according to the manufacturer’s protocol. GO enrichment was analyzed by Metascape with the default setting (https://metascape.org/) (55).
Analysis of published scRNA-seq data.
To validate our cell clustering data obtained with 10× Visium, we analyzed previously published scRNA-seq data sets (15) for day 4 mouse uteri (3.5 dpc in the original paper). Raw single-end RNA-seq reads were aligned to the indexed mouse genome (GRCm38/mm10) using DropSeqTools (https://github.com/broadinstitute/Drop-seq) with default settings. Reanalyses of scRNA-seq data were conducted using the Seurat package for R (v.5.1.0) (56).
3D visualization of implantation sites.
3D visualization of day 6 implantation sites was performed as previously reported (29). To stain luminal and glandular epithelial cells, day 6 tissues were incubated with anti–E-cadherin antibodies (24E10, Cell Signaling Technology; 1:500) followed by incubation with an anti-rabbit antibody conjugated with Alexa Fluor 555 (A21428, Thermo Fisher Scientific; 1:500). 3D images were acquired using LSM 880 (Zeiss) and AXR (Nikon) microscopes. To construct a 3D structure from the images, the surface tool in Imaris (version 9.8, Oxford Instruments) was used.
Measurement of serum P4 levels.
Blood samples were collected from mice on the indicated days of pregnancy. Serum P4 levels were measured as described previously (26), using a progesterone EIA kit (582601, Cayman).
Statistics.
Statistical analyses were performed using a 2-tailed Student’s t test or 1-way ANOVA followed by Bonferroni’s post hoc test in GraphPad Prism 10. Statistical significance was set at a P value of less than 0.05.
Study approval.
All animal experiments were approved by the Institutional Animal Experiment Committee of the University of Tokyo Graduate School of Medicine (approval numbers P16-066, P20-076, and A2023M165).
Data availability.
RNA-seq data reported in this study were deposited to the NCBI Gene Expression Omnibus (GEO accession no. GSE253520). Values for all data points in the graphs are provided in the supplemental Supporting Data Values file.
Author contributions
S Aikawa, MM, S Akaeda, and YH designed the study. S Aikawa, MM, S Akaeda, MA, YI, Y Sugiura, ST, RM, DH, XH, CI, RI, YF, and TH performed the experiments and collected the data. S Aikawa, MM, S Akaeda, MA, YI, Y Sugiura, and YH analyzed the data. Y Sugimoto, RSH, NT, MH, OWH, and YO discussed and interpreted the results. S Aikawa drafted the manuscript, which was edited by YH and Y Sugimoto. RSH critically reviewed the manuscript. YH supervised the study.
Supplementary Material
Acknowledgments
We thank Harvey R. Herschman for providing Ptgs2-floxed mice, Francesco J. DeMayo and John P. Lydon for providing Pgr-Cre mice, and Atsumi Miura for technical assistance. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant nos. JP23K27176, JP23K08278, JP23K15827, JP23K23803, JP23K24481, JP24K22157, JP24K21911, JP24K23524, and JP20H00495), Japan Agency for Medical Research and Development (AMED) (grant nos. JP23gk0210028, JP24gk0210039, JP24gn0110085, JP24gn0110069, and JP24lk0310083), Children and Families Agency (grant no. JPMH23DB0101), Japan Science and Technology Agency (JST) Fusion Oriented Research for disruptive Science and Technology (FOREST) (grant no. JPMJFR210H), Mochida Memorial Foundation for Medical and Pharmaceutical Research, Inoue Research Award for Young Scientists, Uehara Memorial Foundation, Astellas Foundation for Research on Metabolic Disorders, and funds from joint research with NIPRO Corporation.
Version 1. 10/08/2024
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2024, Aikawa et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2024;9(19):e181865.https://doi.org/10.1172/jci.insight.181865.
Contributor Information
Shizu Aikawa, Email: s.aikawa0709@gmail.com.
Mitsunori Matsuo, Email: machio1128@hotmail.co.jp.
Shun Akaeda, Email: shakaeda@gmail.com.
Yukihiko Sugimoto, Email: ysugi@kumamoto-u.ac.jp.
Makoto Arita, Email: makoto.arita@riken.jp.
Yosuke Isobe, Email: yosuke.isobe@riken.jp.
Yuki Sugiura, Email: yuki.sgi@gmail.com.
Shu Taira, Email: taira@agri.fukushima-u.ac.jp.
Rae Maeda, Email: maeda.rei.8n@kyoto-u.ac.jp.
Ryoko Shimizu-Hirota, Email: rshimizu@keio.jp.
Norihiko Takeda, Email: ntakeda-tky@umin.ac.jp.
Daiki Hiratsuka, Email: hiratsuka.daiki@gmail.com.
Xueting He, Email: hexueting0106@outlook.com.
Chihiro Ishizawa, Email: chihiro199191@gmail.com.
Rei Iida, Email: reiiida10888@yahoo.co.jp.
Yamato Fukui, Email: fukuiyamato@gmail.com.
Takehiro Hiraoka, Email: t_hiraoka1985@yahoo.co.jp.
Miyuki Harada, Email: haradam-tky@umin.ac.jp.
Osamu Wada-Hiraike, Email: osamuwh-tky@umin.ac.jp.
Yutaka Osuga, Email: yutakaos-tky@umin.ac.jp.
Yasushi Hirota, Email: yhirota-tky@umin.ac.jp.
References
- 1.Cha J, et al. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18(12):1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukui Y, et al. Uterine receptivity, embryo attachment, and embryo invasion: multistep processes in embryo implantation. Reprod Med Biol. 2019;18(3):234–240. doi: 10.1002/rmb2.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hama K, et al. Embryo spacing and implantation timing are differentially regulated by LPA3-mediated lysophosphatidic acid signaling in mice. Biol Reprod. 2007;77(6):954–959. doi: 10.1095/biolreprod.107.060293. [DOI] [PubMed] [Google Scholar]
- 4.Sugimoto Y, et al. Roles of prostaglandin receptors in female reproduction. J Biochem. 2015;157(2):73–80. doi: 10.1093/jb/mvu081. [DOI] [PubMed] [Google Scholar]
- 5.Dubois RN, et al. Cyclooxygenase in biology and disease. FASEB J. 1998;12(12):1063–1073. doi: 10.1096/fasebj.12.12.1063. [DOI] [PubMed] [Google Scholar]
- 6.Aikawa S, Hirota Y. Roles of lipid mediators in early pregnancy events. Reprod Med Biol. 2024;23(1):e12597. doi: 10.1002/rmb2.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami M, Kudo I. Recent advances in molecular biology and physiology of the prostaglandin E2-biosynthetic pathway. Prog Lipid Res. 2004;43(1):3–35. doi: 10.1016/S0163-7827(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 8.Song H, et al. Cytosolic phospholipase A2alpha is crucial [correction of A2alpha deficiency is crucial] for ‘on-time’ embryo implantation that directs subsequent development. Development. 2002;129(12):2879–2889. doi: 10.1242/dev.129.12.2879. [DOI] [PubMed] [Google Scholar]
- 9.Reese J, et al. Coordinated regulation of fetal and maternal prostaglandins directs successful birth and postnatal adaptation in the mouse. Proc Natl Acad Sci U S A. 2000;97(17):9759–9764. doi: 10.1073/pnas.97.17.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross GA, et al. Opposing actions of prostaglandins and oxytocin determine the onset of murine labor. Proc Natl Acad Sci U S A. 1998;95(20):11875–11879. doi: 10.1073/pnas.95.20.11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim H, et al. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell. 1997;91(2):197–208. doi: 10.1016/S0092-8674(00)80402-X. [DOI] [PubMed] [Google Scholar]
- 12.Akaeda S, et al. Retinoblastoma protein promotes uterine epithelial cell cycle arrest and necroptosis for embryo invasion. EMBO Rep. 2021;22(2):e50927. doi: 10.15252/embr.202050927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, et al. Entosis allows timely elimination of the luminal epithelial barrier for embryo implantation. Cell Rep. 2015;11(3):358–365. doi: 10.1016/j.celrep.2015.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirota Y. Progesterone governs endometrial proliferation-differentiation switching and blastocyst implantation. Endocr J. 2019;66(3):199–206. doi: 10.1507/endocrj.EJ18-0431. [DOI] [PubMed] [Google Scholar]
- 15.Wang HQ, et al. Maternal and embryonic signals cause functional differentiation of luminal epithelial cells and receptivity establishment. Dev Cell. 2023;58(21):2376–2392. doi: 10.1016/j.devcel.2023.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Hirai H, et al. Prostaglandin D2 selectively induces chemotaxis in T helper type 2 cells, eosinophils, and basophils via seven-transmembrane receptor CRTH2. J Exp Med. 2001;193(2):255–261. doi: 10.1084/jem.193.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forman BM, et al. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci U S A. 1997;94(9):4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim H, Dey SK. PPAR delta functions as a prostacyclin receptor in blastocyst implantation. Trends Endocrinol Metab. 2000;11(4):137–142. doi: 10.1016/S1043-2760(00)00243-5. [DOI] [PubMed] [Google Scholar]
- 19.Hara S. Prostaglandin terminal synthases as novel therapeutic targets. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(9):703–723. doi: 10.2183/pjab.93.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paria BC, et al. Zonula occludens-1 and E-cadherin are coordinately expressed in the mouse uterus with the initiation of implantation and decidualization. Dev Biol. 1999;208(2):488–501. doi: 10.1006/dbio.1999.9206. [DOI] [PubMed] [Google Scholar]
- 21.Yuan J, et al. Primary decidual zone formation requires Scribble for pregnancy success in mice. Nat Commun. 2019;10(1):5425. doi: 10.1038/s41467-019-13489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goolam M, et al. The transcriptional repressor Blimp1/PRDM1 regulates the maternal decidual response in mice. Nat Commun. 2020;11(1):2782. doi: 10.1038/s41467-020-16603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Psychoyos A. Endocrine control of egg implantation. In: Greep RO, Astwood EB, eds. Hanbook of Physiology. American Physiology Society; 1973:187–215. [Google Scholar]
- 24.Hirota Y, et al. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest. 2010;120(3):803–815. doi: 10.1172/JCI40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukui Y, et al. The EZH2-PRC2-H3K27me3 axis governs the endometrial cell cycle and differentiation for blastocyst invasion. Cell Death Dis. 2023;14(5):320. doi: 10.1038/s41419-023-05832-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto L, et al. HIF2α in the uterine stroma permits embryo invasion and luminal epithelium detachment. J Clin Invest. 2018;128(7):3186–3197. doi: 10.1172/JCI98931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aikawa S, et al. Uterine deficiency of high-mobility group box-1 (HMGB1) protein causes implantation defects and adverse pregnancy outcomes. Cell Death Differ. 2019;27(5):1489–1504. doi: 10.1038/s41418-019-0429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arora R, et al. Insights from imaging the implanting embryo and the uterine environment in three dimensions. Development. 2016;143(24):4749–4754. doi: 10.1242/dev.144386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J, et al. Tridimensional visualization reveals direct communication between the embryo and glands critical for implantation. Nat Commun. 2018;9(1):603. doi: 10.1038/s41467-018-03092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukui Y, et al. Uterine epithelial LIF receptors contribute to implantation chamber formation in blastocyst attachment. Endocrinology. 2021;162(11):bqab169. doi: 10.1210/endocr/bqab169. [DOI] [PubMed] [Google Scholar]
- 31.Flores D, et al. Mechanical and signaling mechanisms that guide pre-implantation embryo movement. Development. 2020;147(24):dev193490. doi: 10.1242/dev.193490. [DOI] [PubMed] [Google Scholar]
- 32.Ye X, et al. LPA3-mediated lysophosphatidic acid signalling in embryo implantation and spacing. Nature. 2005;435(7038):104–108. doi: 10.1038/nature03505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, et al. Mice missing Cnr1 and Cnr2 show implantation defects. Endocrinology. 2019;160(4):938–946. doi: 10.1210/en.2019-00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenbaum S, et al. A spatially resolved timeline of the human maternal-fetal interface. Nature. 2023;619(7970):595–605. doi: 10.1038/s41586-023-06298-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arutyunyan A, et al. Spatial multiomics map of trophoblast development in early pregnancy. Nature. 2023;616(7955):143–151. doi: 10.1038/s41586-023-05869-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, et al. Spatial transcriptomic profiles of mouse uterine microenvironments at pregnancy day 7.5†. Biol Reprod. 2022;107(2):529–545. doi: 10.1093/biolre/ioac061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakraborty I, et al. Developmental expression of the cyclo-oxygenase-1 and cyclo-oxygenase-2 genes in the peri-implantation mouse uterus and their differential regulation by the blastocyst and ovarian steroids. J Mol Endocrinol. 1996;16(2):107–122. doi: 10.1677/jme.0.0160107. [DOI] [PubMed] [Google Scholar]
- 38.Li X, et al. Isoform-specific compensation of cyclooxygenase (Ptgs) genes during implantation and late-stage pregnancy. Sci Rep. 2018;8(1):12097. doi: 10.1038/s41598-018-30636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kudo K, et al. Secreted phospholipase A2 modifies extracellular vesicles and accelerates B cell lymphoma. Cell Metab. 2022;34(4):615–633. doi: 10.1016/j.cmet.2022.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Chen Q, et al. Transient {beta}2-adrenoceptor activation confers pregnancy loss by disrupting embryo spacing at implantation. J Biol Chem. 2011;286(6):4349–4356. doi: 10.1074/jbc.M110.197202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dawson M, et al. Imaging the dynamics of murine uterine contractions in early pregnancy†. Biol Reprod. 2024;110(6):1175–1190. doi: 10.1093/biolre/ioae071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsumoto H, et al. Cyclooxygenase-2 differentially directs uterine angiogenesis during implantation in mice. J Biol Chem. 2002;277(32):29260–29267. doi: 10.1074/jbc.M203996200. [DOI] [PubMed] [Google Scholar]
- 43.Deng W, et al. Endothelial cells in the decidual bed are potential therapeutic targets for preterm birth prevention. Cell Rep. 2019;27(6):1755–1768. doi: 10.1016/j.celrep.2019.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruan YC, et al. Activation of the epithelial Na+ channel triggers prostaglandin E2 release and production required for embryo implantation. Nat Med. 2012;18(7):1112–1117. doi: 10.1038/nm.2771. [DOI] [PubMed] [Google Scholar]
- 45.Vinketova K, et al. Human decidual stromal cells as a component of the implantation niche and a modulator of maternal immunity. J Pregnancy. 2016;2016:8689436. doi: 10.1155/2016/8689436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishihara E, et al. Prostaglandin E2 and its receptor EP2 trigger signaling that contributes to YAP-mediated cell competition. Genes Cells. 2020;25(3):197–214. doi: 10.1111/gtc.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marions L, Danielsson KG. Expression of cyclo-oxygenase in human endometrium during the implantation period. Mol Hum Reprod. 1999;5(10):961–965. doi: 10.1093/molehr/5.10.961. [DOI] [PubMed] [Google Scholar]
- 48.Ishikawa TO, Herschman HR. Conditional knockout mouse for tissue-specific disruption of the cyclooxygenase-2 (Cox-2) gene. Genesis. 2006;44(3):143–149. doi: 10.1002/gene.20192. [DOI] [PubMed] [Google Scholar]
- 49.Langenbach R, et al. Prostaglandin synthase 1 gene disruption in mice reduces arachidonic acid-induced inflammation and indomethacin-induced gastric ulceration. Cell. 1995;83(3):483–492. doi: 10.1016/0092-8674(95)90126-4. [DOI] [PubMed] [Google Scholar]
- 50.Soyal SM, et al. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis. 2005;41(2):58–66. doi: 10.1002/gene.20098. [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto H, et al. Molecular and cellular events involved in the completion of blastocyst implantation. Reprod Med Biol. 2016;15(2):53–58. doi: 10.1007/s12522-015-0222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Dewar A, et al. Tissue-specific RNA localization in the uterus during implantation, decidualization and placentation: technical nuances of various labeling approaches. Curr Protoc. 2023;3(6):e823. doi: 10.1002/cpz1.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aikawa S, et al. Autotaxin-lysophosphatidic acid-LPA3 signaling at the embryo-epithelial boundary controls decidualization pathways. EMBO J. 2017;36(14):2146–2160. doi: 10.15252/embj.201696290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data reported in this study were deposited to the NCBI Gene Expression Omnibus (GEO accession no. GSE253520). Values for all data points in the graphs are provided in the supplemental Supporting Data Values file.