FIGURE.
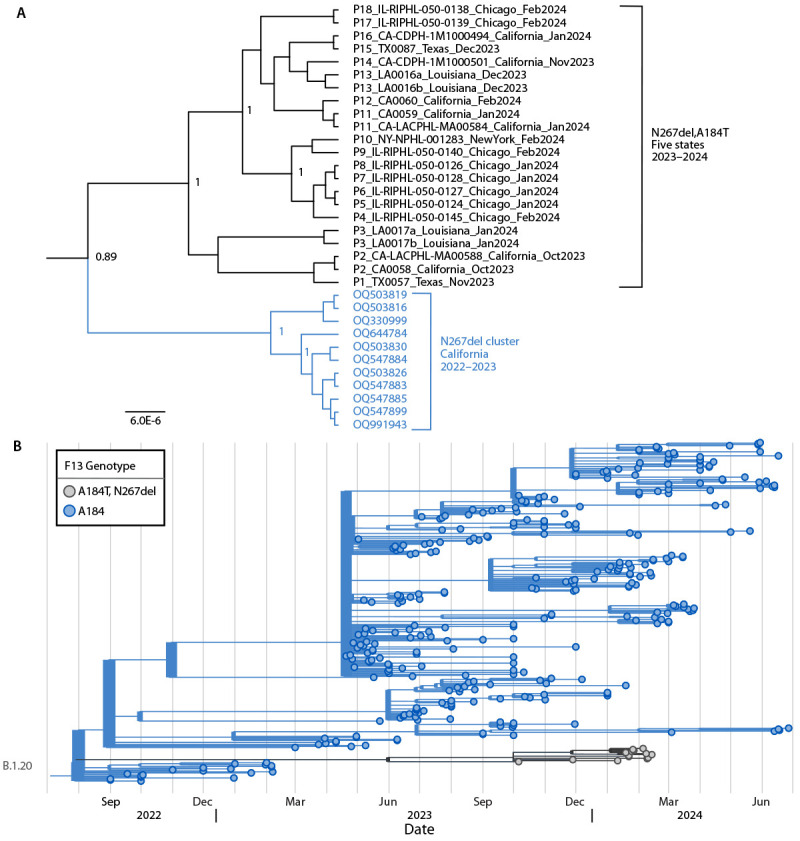
Phylogeny showing the relatedness of tecovirimat-resistant monkeypox viruses among 18 U.S. cases during 2023–2024*,† with a previous tecovirimat-resistant cluster from California (A)§,¶ and with representative monkeypox virus sequences (B)** — United States, 2023–2024
Abbreviations: MPXV = monkeypox virus; A184T = alanine-184-to-threonine substitution; N267del = asparagine 267 deletion; SNP = single nucleotide polymorphism.
* From five states (California, Illinois, Louisiana, New York, and Texas).
† Unique patients are identified by P number; multiple samples were sequenced for four patients.
§ MPXV with N267del, A184T clustered separately from MPXV sequences from a tecovirimat-resistant cluster from California with N267del from late 2022–early 2023 (sublineage B.1.17). During late 2022–early 2023, persons in two of the four California cases (P16 and P2) reported travel to Texas; each had one or two SNP differences from an MPXV sequence from Texas (P15 and P1, respectively). Within the cluster, N267del, A184T sequences differed by an average of five SNPs (range = 0–11), compared with an average of 10.5 SNPs between the two clusters (range = 7–15). A large genomic deletion in sequence TX0087 was counted as one SNP.
¶ Phylogenetic analysis was performed using Bayesian Evolutionary Analysis Sampling Trees (BEAST; version 1.10.4) on whole genome sequence alignments of sequences after clipping genomic ends to match the shortest sequence, removing repeat regions, and removing any sites containing ambiguous bases to a final length of 150,505 positions using GTR+ G+I nucleotide substitution model, exponential coalescent tree prior, and strict molecular clock set to one; ON676708 was used to root the tree. Scale bar indicates substitutions per site; numbers at the branches indicate posterior probability support values.
** MPXV with N267del, A184T clustered separately from other sublineage B.1.20 MPXV sequences. Visualization of 334 lineage B.1.20 clade IIb MPXV genomes sampled from 3,552 genomes available from the National Center for Biotechnology Information GenBank between September 2022 and June 2024 (updated July 22, 2024) using a local build of Nextstrain (https://nextstrain.org) and visualized using TimeTree (https://timetree.org).
