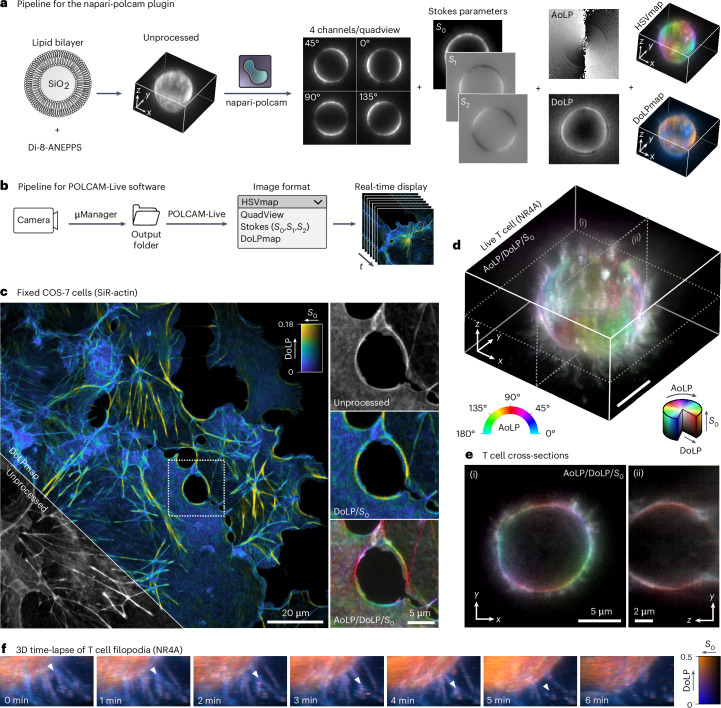
Fig. 6. Diffraction-limited polarization microscopy using POLCAM.
a, Image processing workflow of the napari plugin napari-polcam, demonstrated on an example 3D dataset of a lipid bilayer-coated silica microsphere labeled with the membrane dye Di-8-ANEPPS. b, Workflow of POLCAM-Live software for real-time processing and rendering of polarization camera images during image acquisition. c, Diffraction-limited polarization camera image of fixed COS-7 cells labeled with SiR–actin, rendered using a DoLP color map. An inset marked by a dotted square is shown in an unprocessed, DoLP color map and an HSV polarization color map. d, A 3D image of the plasma membrane (using the dye NR4A) of a live T cell. e, Two cross-sections of the T cell shown in d. f, A 3D time-lapse of the movement of the filopodia of a live T cell rendered using a DoLP color map. Randomly polarized regions appear blue (filopodia) and more structured, and therefore polarized areas (larger, more smooth sections of the plasma membrane surface) appear orange. A white triangle tracks the movement of what appears to be a branching point on a filopodium.