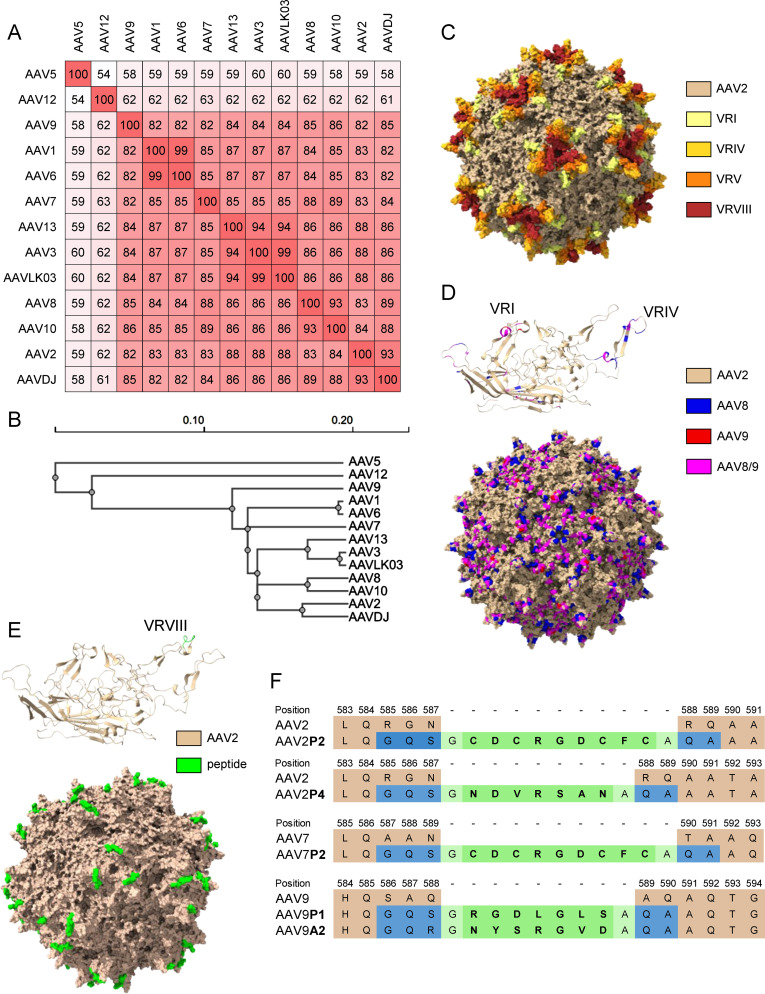
Figure 1.
Sequence information for the AAV capsids used in this study. (A) VP1 amino acid sequence percentage identity matrix for all wild-type and shuffled AAV capsids used in this study and (B) guide tree of the same set of AAVs generated using Clustal Omega 1.2.4. (C) Visual representation of the AAV2 capsid structure [PDB ID 6IH9; (38)], the variable regions VRI, VRIV, VRV and VRVIII are highlighted in light yellow, yellow, orange and red, respectively. (D) Visual representation of the AAVDJ VP1 capsid protein (top) and whole capsid structure [bottom; PDBID: 7KFRM; (39)] with sequences derived from AAV2, AAV8 and AAV9 indicated by color. Wheat, blue and red indicate sequences from AAV2, AAV8 and AAV9, respectively, while magenta represents sequences that are identical in both AAV8 and AAV9. The labelled VRs in the VP1 structure represent the loops that were particularly divergent from AAV2. (E) Visual representation of peptide-modified capsids using the AAV2P2 VP1 capsid protein monomer (top) and the assembled capsid (bottom) as a representative example of the peptide-modified AAVs used in this study. The AAV2 sequence is shown as wheat colored, the inserted peptide sequence is colored in green. (F) Amino acid sequences surrounding the peptide insertion site in the peptide-modified AAV capsids used in this study. Wheat colored amino acids are the parental sequences, green colored amino acids are the inserted peptides. Blue colored amino acids resulted from the introduction of restriction enzyme cleavage sites to accommodate the peptide-encoding oligonucleotides, light green colored amino acids indicate glycine and alanine linker residues flanking all inserted peptide sequences.