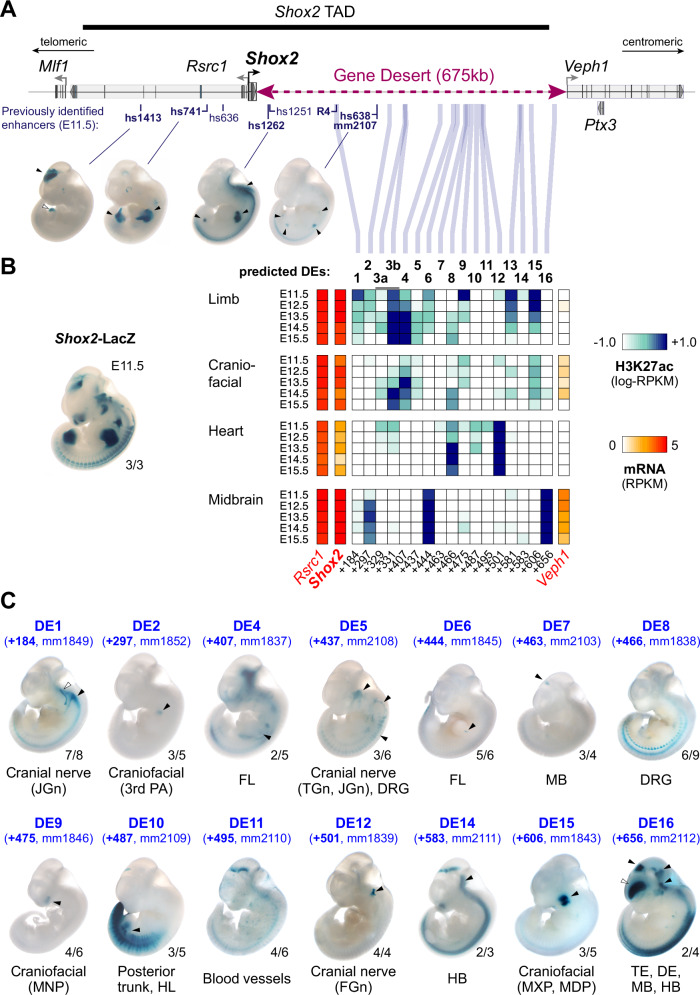
Fig. 1. The Shox2 gene desert constitutes a hub for tissue-specific enhancers.
A Genomic interval containing the Shox2 TAD64 and previously identified Shox2-associated enhancer regions. Vista Enhancer Browser IDs (hs: human sequence, mm: mouse sequence) in bold mark enhancers with Shox2-overlapping and reproducible activities (arrowheads). The position of the human R4 enhancer55 driving reporter activity in the sinus venosus is indicated. B Heatmap showing H3K27 acetylation (ac) -predicted and ChromHMM-filtered putative enhancers and their temporal signatures in tissues with dominant Shox2 functions (see full Supplementary Fig. 1). Blue and red shades represent H3K27ac enrichment and mRNA expression levels, respectively. Distance to Shox2 TSS (+) is indicated in kb. Left: Shox2 expression pattern (Shox2-LacZ/+) at E11.532. C Transgenic LacZ reporter validation of predicted gene desert enhancers (DEs) in mouse embryos at E11.5. Arrowheads point to reproducible enhancer activity with (black) or without (white) Shox2 overlap. JGn, TGn, FGn: jugular, trigeminal, and facial ganglion, respectively. PA, pharyngeal arch. DRG, dorsal root ganglia. FL, Forelimb. HL, Hindlimb. TE, Telencephalon. DiE, Diencephalon. MB, Midbrain. HB, Hindbrain. MNP, medial nasal process. MXP, maxillary process. MDP, mandibular process. Reproducibility numbers are indicated on the bottom right of each representative embryo shown (reproducible tissue-specific staining vs. number of transgenic embryos with any LacZ signal). Corresponding Vista IDs of the elements tested are listed in Supplementary Table 1.