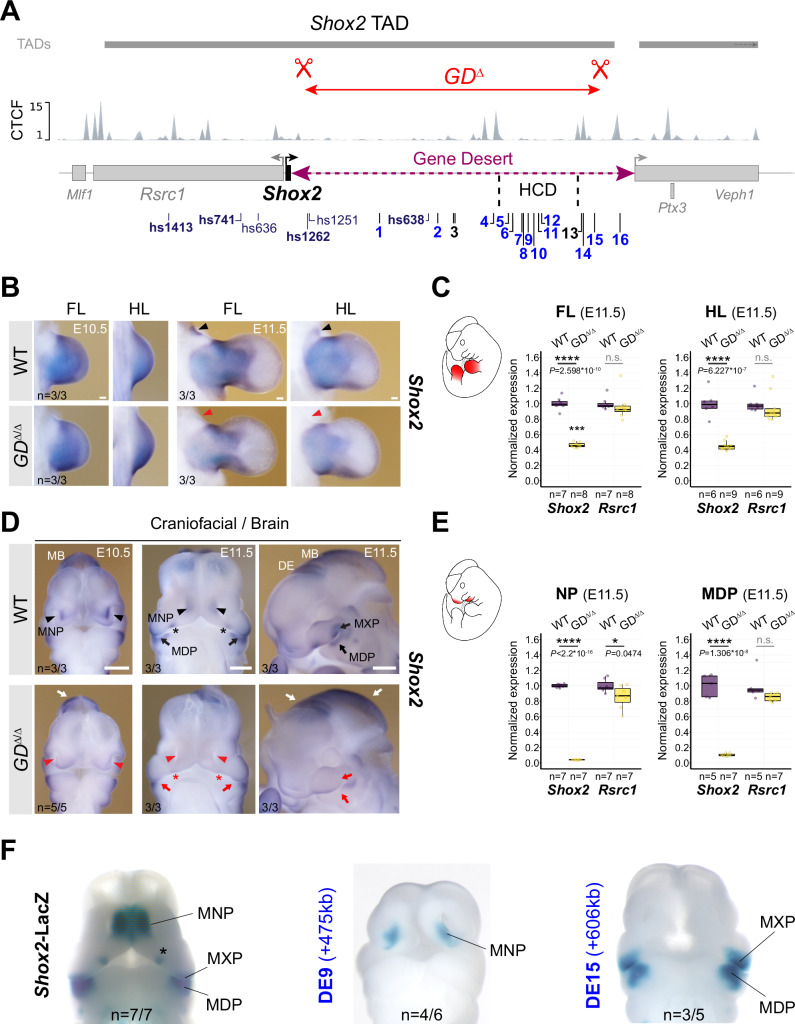
Fig. 3. Gene desert deletion reduces Shox2 in limb and craniofacial compartments.
A CRISPR/Cas9-mediated deletion of the intra-TAD Shox2 gene desert interval (GDΔ) (mm10, chr3:66365062-66947168). Vista (hs) and newly identified gene desert enhancers (1-16, active in blue) are displayed along with TAD interval and CTCF peaks from mESCs64. HCD, high-density contact domain (see Fig. 2). B, D ISH revealing spatial Shox2 expression in fore- and hindlimb (FL/HL), craniofacial compartments, and brain in GDΔ/Δ embryos compared to wildtype (WT) controls at E10.5 and E11.5. Red arrowheads and red arrows point to regions with severely downregulated or reduced Shox2 expression, respectively. Red asterisk demarcates Shox2 loss in the anterior portion of the palatal shelves. White arrows indicate regions (diencephalon, DE and midbrain, MB) without overt changes in Shox2 expression. Scale bars, 500 μm (b) and 100 μm (d). C, E Quantitative mRNA analysis (qPCR) in limb and craniofacial tissues of WT and GDΔ/Δ embryos. Box plots indicate interquartile range, median, maximum/minimum values (bars). Dots represent individual data points. ****P < 0.0001; *P < 0.05; n.s., not significant (two-tailed, unpaired t-test for qPCR). F DE9 and DE15 enhancer activities (Fig. 1C) overlap Shox2 expression in medial nasal process (MNP) and maxillary-mandibular (MXP-MDP) regions, respectively, in mouse embryos at E11.5. Asterisk marks anterior palatal shelf. “n” indicates number of embryos per genotype, or transgene analyzed, with similar results. Source data are provided in the Source Data file.