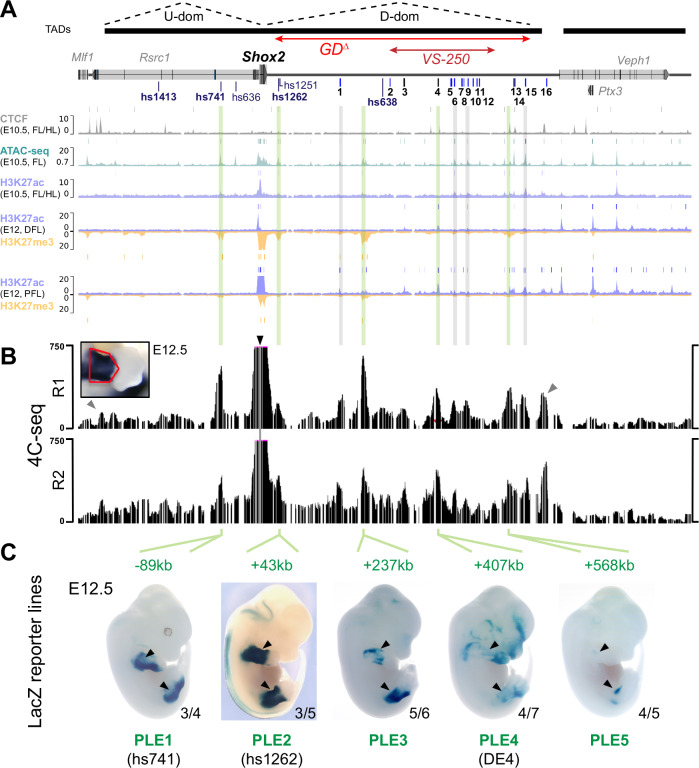
Fig. 6. The gene desert encodes a series of distributed proximal limb enhancers (PLEs) with subregional specificities.
A Re-processed ChIP-seq datasets from mouse embryonic limbs at E10.5 (CTCF, ATAC-seq, H3K27ac) and E12 (H3K27ac, H3K27me3) showing epigenomic profiles at the Shox2 locus59,63,144. Bars above each track represent peak calls across replicates. DFL, distal forelimb. PFL, proximal forelimb. On top: TAD extension in mESCs64 (black bars) with desert enhancers identified in Fig. 1 (DEs 1-16; blue indicates validated activity). The extension of the gene desert deletion (GDΔ) and SV control region (VS-250) is indicated. B 4C-seq interaction profiles from two independent biological replicates (R1, R2) of proximal forelimbs at E12.5 (red outline). Black arrowhead indicates the 4C-seq viewpoint at the Shox2 promoter. Gray arrowheads point to CTCF-boundaries of the Shox2-TAD. Green lines (in A) indicate Shox2-interacting elements with putative proximal limb activities. Gray lines mark 4C-seq peaks overlapping previously validated DEs without such activities (Fig. 1C). C Identification of proximal limb enhancers (PLEs) through transgenic LacZ reporter assays in mouse embryos at E12.5. Embryos shown are representatives from stable transgenic LacZ reporter lines (Supplementary Fig. 6A). Reproducibility numbers from original transgenic founders are listed for each element (bottom right).